Introduction
In wool fibres, the main structural proteins are hard α-keratins. They are assembled in a highly organized fashion into keratin intermediate filaments (KIFs) and enveloped by an inter-filamentous matrix consisting of keratin-associated proteins (KAPs) (Popescu and Höcker, Reference Popescu and Höcker2007). The KIF proteins are typically low-sulphur proteins when compared to the other wool proteins, and they are grouped into type-I and type-II families, according to their site of expression, function and pKa (whether they are acidic or neutral/basic). They can be further subdivided into epithelial cytokeratins and hair keratins (Heid et al., Reference Heid, Werner and Franke1986, Reference Heid, Moll and Franke1988; Lynch et al., Reference Lynch, O'Guin, Hardy, Mak and Sun1986).
The α-keratin K31 belongs to the type I family and is found in the cortex of wool fibres, along with K85 and K38 (Yu et al., Reference Yu, Gordon, Nixon, Bawden, Rogers, Wildermoth, Maqbool and Pearson2009). The K31 gene (KRT31, formerly known as KRT1.1) has been mapped to ovine chromosome 11q25-q29 (Hediger et al., Reference Hediger, Ansari and Stranzinger1991) and is described at ENSOARG00000016473, Oar_v3.1:CM001592.1; Chromosome 11:41 105 175–41 108 801. Eighteen KRTs and KRTAPs have been mapped on the same chromosome near KRT31, and in a cluster (Sumner et al., Reference Sumner, Forrest, Zhou, Henderson and Hickford2013; Gong et al., Reference Gong, Zhou, Forrest, Li, Wang, Dyer, Luo and Hickford2016).
A number of quantitative trait loci (QTLs) and markers for wool traits, including greasy fleece weight (GFW), clean fleece weight (CFW), mean staple strength (MSS) and coefficient of variation of fibre diameter (CVFD) have been reported on ovine chromosome 11 (Rogers et al., Reference Rogers, Hickford and Bickerstaffe1994; Roldan et al., Reference Roldan, Dodero, Bidinost, Taddeo, Allain, Poli and Elsen2010), but there are only a small number of studies describing genetic variation in the chromosome 11 KRT genes.
Rogers et al. (Reference Rogers, Hickford and Bickerstaffe1993) reported a MspI polymorphism in KRT1.2 (now called KRT33a) using a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) approach, and Itenge-Mweza et al. (Reference Itenge-Mweza, Forrest, McKenzie, Hogan, Abbott, Amoafo and Hickford2007) described five alleles of KRT33 (now called KRT33a), using a PCR-single stranded conformational polymorphism (PCR-SSCP) approach. Itenge et al. (Reference Itenge, Hickford, Forrest, McKenzie and Frampton2010) reported that KRTAP1.1, KRTAP1.3 and KRT33a were potential gene markers associated with a variety of wool traits, including MSS, mean staple length (MSL), fibre diameter standard deviation (FDSD), wool yellowness and lustre in Merino and Merino-cross sheep. Genes located within a region near to KRT1.2 could also have an effect on MSS (Parsons et al., Reference Parsons, Piper and Cooper1994; Sumner et al., Reference Sumner, Forrest, Zhou, Henderson and Hickford2013).
In the current study, PCR-SSCP was used to investigate sequence variation in KRT31 and the associations, if any, which may occur between KRT31 sequence variation and various wool traits.
Materials and methods
Sheep and wool samples
There were two parts to the current study. The first was to ascertain the extent of genetic variation in KRT31 in 300 New Zealand (NZ) Merino, Romney and White Dorper sheep, sourced from 26 farms. Four regions of the gene were analysed: a 457-bp fragment of the promoter, a 422-bp fragment spanning the entire exon 1, a 271-bp fragment spanning the entire exon 3 and a 311-bp fragment spanning the entire exon 7 coding sequence.
Next, the association of KRT31 promoter variation with selected wool traits was investigated in a separate group of 485 Southdown × Merino-cross lambs derived from seven sire-lines. These lambs were all managed on the same farm and were shorn at 12 months of age.
At shearing, GFW was measured and wool samples were collected from the mid-side region for wool trait measurement at the New Zealand Wool Testing Authority Ltd (NZWTA, Napier, NZ) using the International Wool Testing Organisation (IWTO) standardized methods. This included measurement of wool yield (Yield; %), MSL (mm), MSS (N/ktex), mean fibre diameter (MFD; μm), FDSD (μm), CVFD (%), mean fibre curvature (MFC; °/mm) and prickle factor (PF; the percentage of fibres of diameter greater than 30 microns). Clean fleece weight (CFW; kg) was calculated from the GFW and Yield measurement.
Polymerase chain reaction amplification
A blood sample from each sheep was collected onto a Whatman Flinders Technology Associates (FTA) card and genomic DNA was purified using a two-step washing procedure as described in Zhou et al. (Reference Zhou, Hickford and Fang2006). The primers used to amplify the four regions of KRT31 were designed based on sequences in the ovine genome assembly v4.0 (Table 1). The primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA).
Table 1. The primer sequences and PCR-SSCP conditions for screening variation in three regions of ovine KRT31
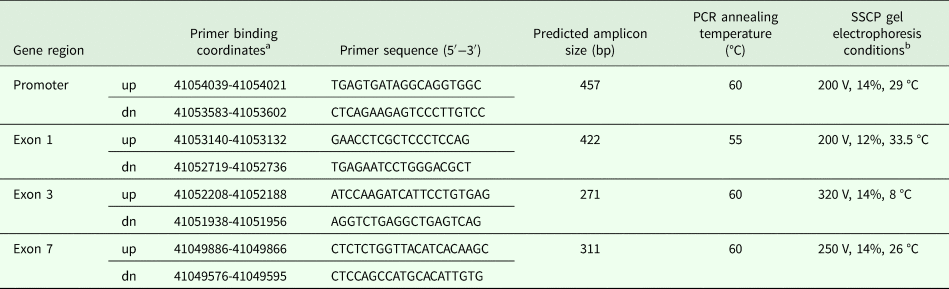
PCR-SSCP, polymerase chain reaction-single stranded conformational polymorphism.
a Refers to the Oar v4.0 sequence NC_019460.
b V, Voltage; %, Acrylamide gel percentage; °C, Electrophoresis temperature.
Polymerase chain reaction amplification was performed in 15-μl reactions and included the genomic DNA on one 1.2 mm punch of FTA paper, 10 × reaction buffer with 0.5 U of Taq DNA polymerase (Qiagen, Hilden, Germany), a 250 nM concentration of each primer, a 150 µM concentration of each deoxyribonucleotide triphosphate (dNTP; Eppendorf, Hamburg, Germany) and a final magnesium ion (Mg2+) concentration of 2.5 mM. The cycling parameters for PCR amplification consisted of denaturation at 94 °C for 2 min., followed by 35 cycles of 30 s each of denaturation at 95 °C, annealing at the temperature shown in Table 1 and extension at 72 °C; with a final extension at 72 °C for 5 min. The amplifications were undertaken in either iCyclers (Bio-Rad, Hercules, CA, USA) or S1000 thermal cyclers (Bio-Rad).
Variation screening and DNA sequencing
All SSCP analyses were carried out in 14% or 12% polyacrylamide gels (37.5:1; Bio-Rad) in 0.5 × Tris/Borate/Ethylenediaminetetraacetic acid (TBE) buffer under the electrophoresis conditions described in Table 1 for 16 h. A 0.7-μl aliquot of the product from the PCR amplification was mixed with 7 µl of loading dye (98% formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene-cyanol), denatured at 95 °C for 5 min, then cooled rapidly on wet ice and loaded onto gels. All gels were silver-stained according to the method of Byun et al. (Reference Byun, Fang, Zhou and Hickford2009).
Polymerase chain reaction amplicons from sheep homozygous for each of the representative SSCP banding patterns were sequenced directly at the Lincoln University DNA Sequencing Facility, Lincoln, New Zealand. For variants that were only found in apparently heterozygous sheep, the DNA was sequenced using an approach described in Gong et al. (Reference Gong, Zhou, Yu, Dyer, Plowman and Hickford2011). Briefly, the SSCP band corresponding to the variant not found in a homozygous form was excised from the SSCP gel, macerated and used as a template for re-amplification. The product of the second amplification was then directly sequenced in triplicate.
Sequence alignments, translations (to determine presumed amino acid sequences) and comparisons were carried out using DNAMAN (version 5.2.10, LynnonBioSoft, Vaudreuil, Canada). The Basic Local Alignment Search Tool (BLAST) algorithm was used to search the National Center for Biotechnology Information (NCBI) GenBank (http://www.ncbi.nlm.nih.gov/) databases for sequence homology, in particular, homology found with the ovine genome assembly v4.0.
Genotyping of the promoter region
Following initial screening of the 300 NZ Romney, Merino and White Dorper sheep, an inner reverse PCR primer (5′-GTTAGTCCATCCAATGACATTC-3′) for the KRT31 promoter region was designed to amplify a smaller fragment that spanned the nucleotide variations identified with the first set of primers. This primer was used along with the reverse primer at an annealing temperature of 60 °C, to type the KRT31 promoter region in the 489 Southdown × Merino-cross lambs using PCR-SSCP in 14% polyacrylamide gels (37.5:1; Bio-Rad) under the electrophoresis conditions of 200 V, 29 °C for 16 h. The reference samples for each variant were included in each gel to facilitate precise genotyping.
Statistical analysis
For each wool trait, statistical analyses were performed using Minitab version 16. Unless otherwise indicated, all P-values were considered statistically significant when P < 0.05 and trends were noted when 0.05 ⩽ P < 0.10.
General Linear Mixed-effects Models (GLMMs) were used to evaluate the effect, if any, of the presence/absence of the KRT31 promoter variants, or KRT31 genotypes, on various wool traits. In all the models, gender and birth rank were fitted as fixed factors, and sire was fitted as a random factor.
The KRT31 promoter variants were coded as either present (1) or absent (0) for each animal's genotype. For each wool trait, single-variant presence/absence models were then run. Any gene variant that had an association in the single-variant models with a P-value <0.2, and which could thus potentially impact on the trait, was then factored into multi-variant models, such that a correction was made for this variant in the genotype.
In the third set of models, any KRT31 promoter genotypes present at a frequency of 5% or more (thereby ensuring adequate sample size) were tested to ascertain associations with the wool traits. Multiple pairwise comparisons between genotypes were performed using a Tukey test with Bonferroni corrections.
Results
Variants of the KRT31 promoter, exon 1, exon 3 and exon 7 regions
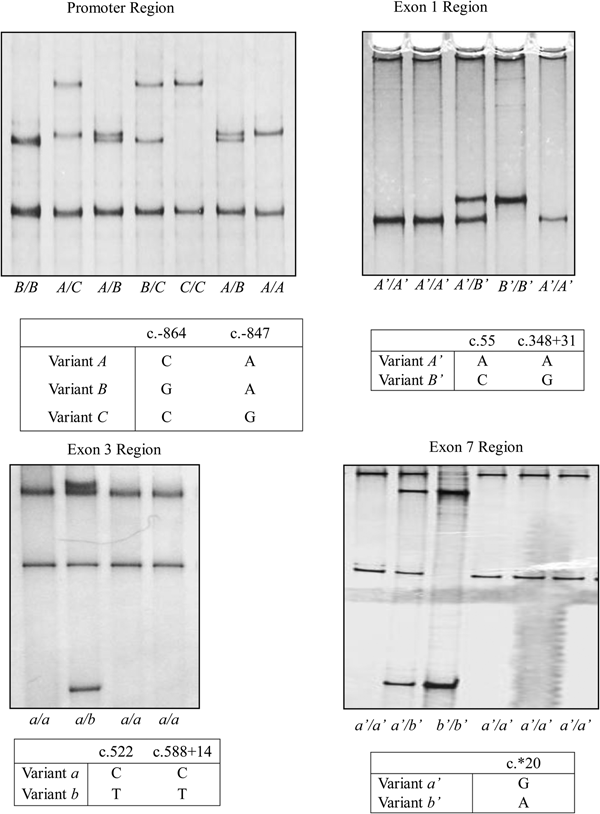
For the regions investigated (KRT31 promoter, exon 1, exon 3 and exon 7), three, two, two and two PCR-SSCP banding patterns were observed, respectively. Sequencing of the amplicons that produced unique SSCP banding patterns for each of the four regions of KRT31 revealed sequences that were unique, but at least 99% homologous to the ovine genome (ovine genome assembly v4.0, 41053583-41054040, 41052719-41053137, 41051938-41052208, 41049576-41049866; for the KRT31 promoter, exon 1, exon 3 and exon 7 regions, respectively).
There were two single nucleotide polymorphisms (SNPs) identified in the KRT31 promoter region from the 300 NZ Romney, Merino and White Dorper sheep typed: c.−864C/G and c.−847A/G (Fig. 1). These gave rise to three variants (designated A, B and C; Fig. 1). Two nucleotide sequences were confirmed for the KRT31 exon 1 region amplified in the 300 NZ Romney, Merino and White Dorper sheep and these were designated A’ and B’ (Fig. 1). They contained two SNPs: a synonymous SNP c.55A/C and an intronic SNP c.348+31A/G. The exon 3 region that was amplified produced two nucleotide sequences (designated a and b), and two SNPs c.522C/T, c.588+14C/T were detected (Fig. 1). The nucleotide substitution in the coding region was synonymous, and the other was in intron 3. Two unique variants were observed for the exon 7 region amplified and these were named a’ and b’. They contained a SNP c.*20G/A (Fig. 1), with the nucleotide substitution occurring in a 3′ untranslated region.
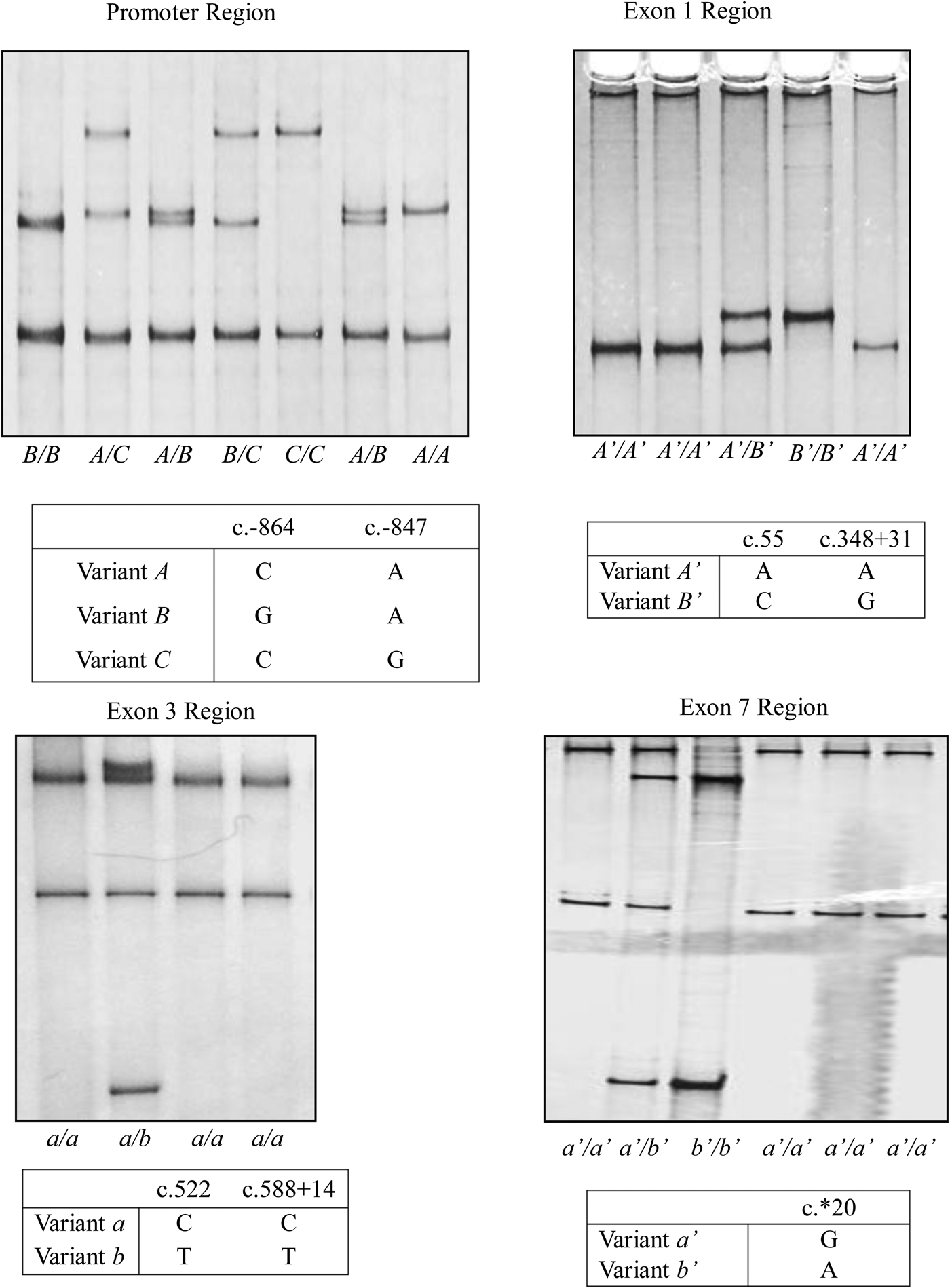
Fig. 1. Variation identified in ovine KRT31. Three PCR-SSCP patterns corresponding to three (A, B and C) variant sequences were identified in the promoter region, while two PCR-SSCP patterns responding to two (A’ and B’) variant sequences were identified in the exon 1 region, two PCR-SSCP patterns responding to two (a and b) variant sequences were identified in the exon 3 region and two PCR-SSCP patterns responding to two (a’ and b’) variant sequences were identified in the exon 7 region. Only the nucleotide sequences that differ between the variants are shown, and nucleotide positions refer to GenBank accession no. NC_019468 following the nomenclature described in http://www.hgvs.org/mutnomen/.
The frequencies of KRT31 promoter variants A to C in the Southdown × Merino-cross sheep were 56, 29 and 15%, respectively. Five different genotypes were observed with the following genotypes having frequencies over 5%: AA (28%); AB (30%); AC (18%); BB (8%) and BC (7%). The remaining genotype, CC, was only observed in 15 sheep.
Associations between variation in the KRT31 promoter and wool traits
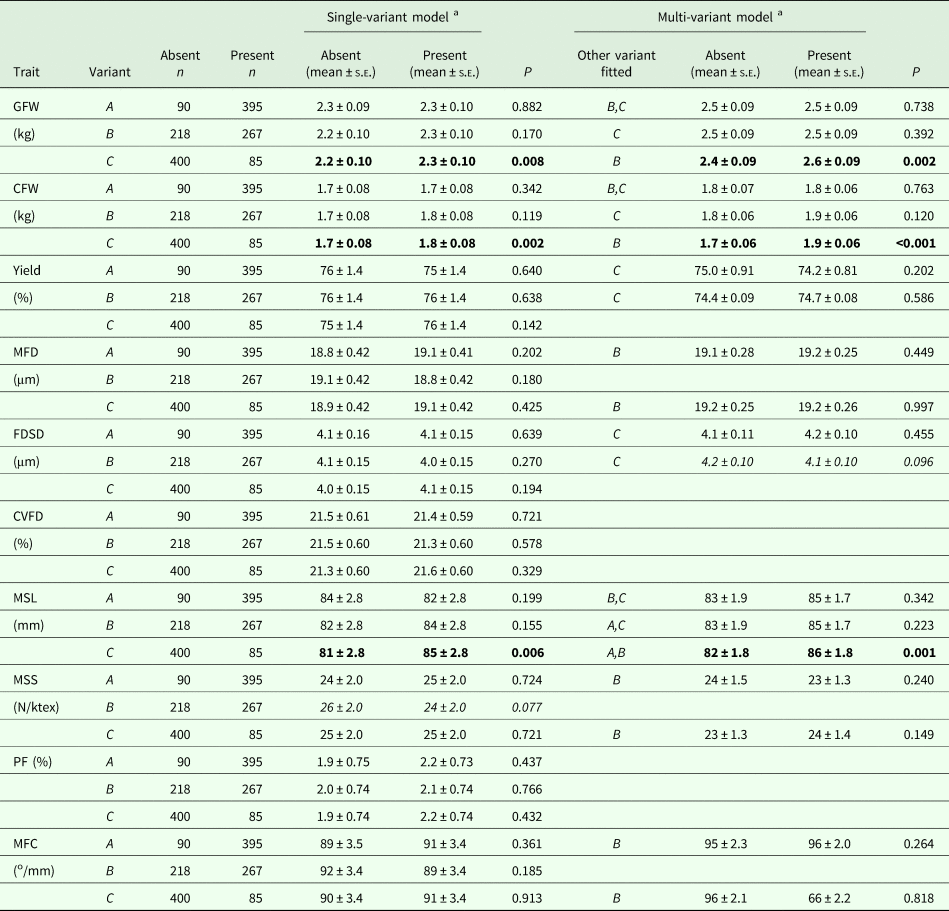
No associations were detected with A and B in the GLMMs, but the presence of C was found to be associated with an increase in GFW, CFW and MSL (Table 2). These effects persisted in the multi-variant GLMMs. The phenotypic change was 5.9%, 7.2% and 4.8%, respectively, for GFW, CFW and MSL.
Table 2. Association of KRT31 promoter variants with various wool traits
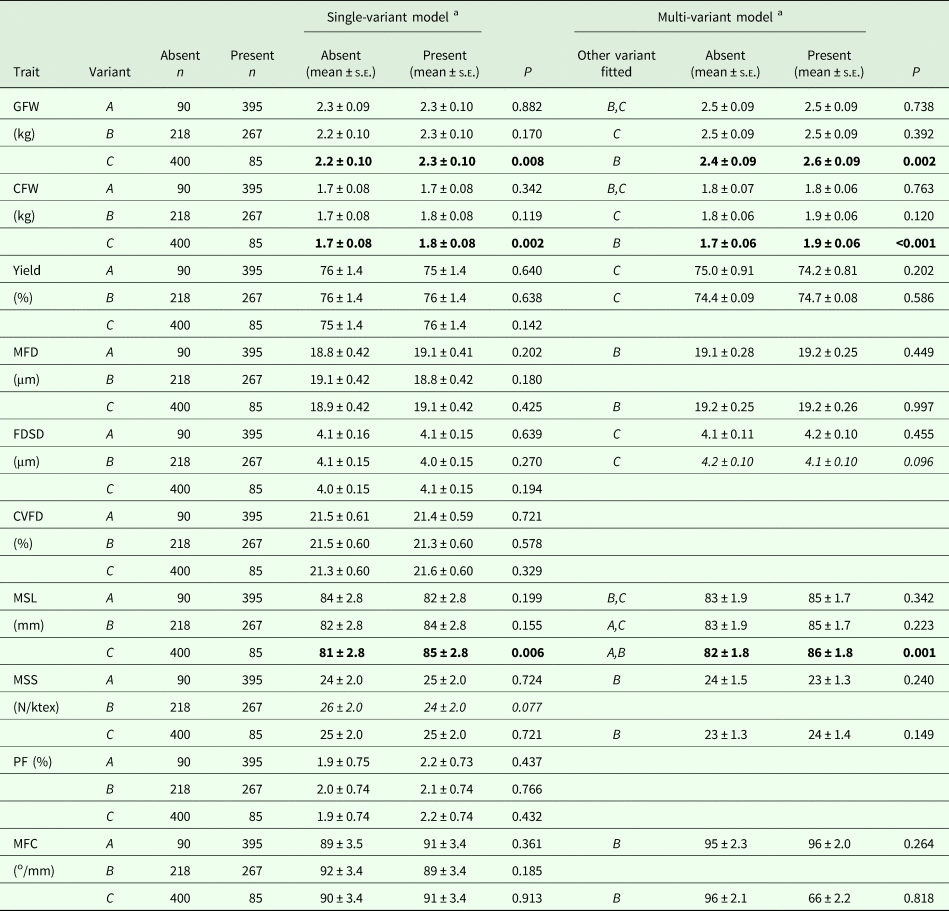
GFW, greasy fleece weight; CFW, clean fleece weight; Yield, wool yield; MFD, mean fibre diameter; FDSD, fibre diameter standard deviation; CVFD, coefficient of variation of fibre diameter; MSL, mean staple length; MSS, mean staple strength; PF, prickle factor (the percentage of fibres of diameter greater than 30 microns); MFC, mean fibre curvature.
a Estimated marginal means and standard errors (s.e.) derived from the GLMMs. P < 0.05 are in bold, whereas 0.05 ⩽P < 0.10 are italicised.
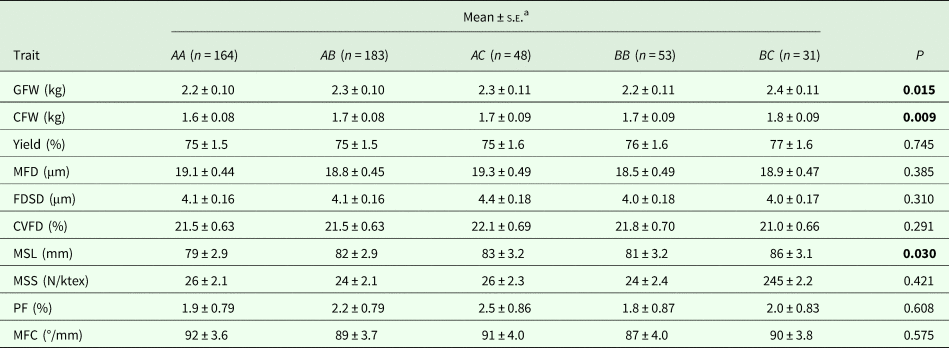
With the five genotypes (AA, AB, AC, BB and BC) that occurred at a frequency >5%, an effect of genotype on GFW, CFW and MSL was detected. Genotype BC sheep produced wool of higher GFW, CFW and MSL than AA, AB, AC and BB sheep (Table 3). Although the overall genotypic effect on GFW was significant (P = 0.015), the pairwise comparison revealed there were no significant pairwise differences in GFW between sheep of differing genotype (Table 3).
Table 3. The effect of KRT31 promoter genotype on various wool traits
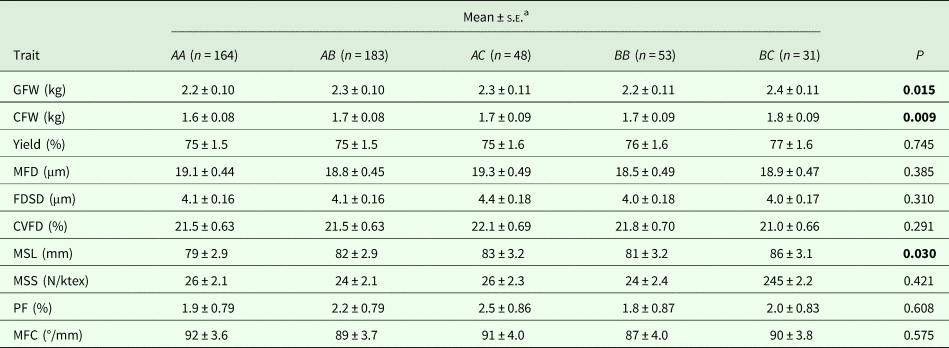
GFW, greasy fleece weight; CFW, clean fleece weight; MFD, mean fibre diameter; FDSD, fibre diameter standard deviation; CVFD, coefficient of variation of fibre diameter; MSL, mean staple length; MSS, mean staple strength; MFC, mean fibre curvature; PF, prickle factor (percentage of fibres greater than 30 microns).
bEstimated marginal means, standard errors (S.E.) and P values derived from GLMs. Bonferroni correction fitted for repetitive testing. P < 0.05 are in bold.
Discussion
Although many of the SNPs detected in KRT31 occurred in introns and would not change the putative amino acid chain produced, variation in the introns of genes can influence gene expression by changing the activity of enhancer elements, or by influencing the production and splicing of primary transcripts (Fong and Zhou, Reference Fong and Zhou2001; Furger et al., Reference Furger, O'Sullivan, Binnie, Lee and Proudfoot2002; Kwek et al., Reference Kwek, Murphy, Furger, Thomas, O'Gorman, Kimura, Proudfoot and Akoulitchev2002). Further analysis of the effect of these SNPs could, therefore, involve precise analysis of the expression level of different variants in sheep of different genotypes.
Three promoter sequence variants were identified and defined by two SNPs (c.−864C/G, and c.−847A/G). Variation in promoters could affect transcription factor binding sites or enhancer elements, which potentially affects the expression of the gene. Keratin promoter regions have proven invaluable for targeting transgene expression to specific compartments within epithelia (Liu et al., Reference Liu, Lyle, Yang and Cotsarelis2003). For instance, the promoter region of KRT15 was found to target hair follicle bulge cells with specificity, and the KRT5 and KRT14 promoters drive expression of transgenes to the relatively undifferentiated basal cell layer of the epidermis and hair follicle (Liu et al., Reference Liu, Lyle, Yang and Cotsarelis2003). It could, therefore, be claimed with some confidence that KRT31 promoter variation might affect gene expression, at least until proven otherwise.
The exon 1, exon 3 and exon 7 sequence variants were identified by two (c.55A/C and c.348+31A/G), two (c.522C/T and c.588+14C/T) and one (c.*20G/A) SNPs, respectively. These SNPs were either synonymous SNPs in the coding region, or located in intron regions, or the 3′ untranslated region. This would not change the putative amino acid sequence but could affect the rate of transcription. The occurrence of SNPs in intron regions can cause inactivation of mRNA splice donor site, which can ultimately result in the formation of premature stop codons or exon skipping, hence yielding a shorter mRNA (Kimchi-Sarfaty et al., Reference Kimchi-Sarfaty, Oh, Kim, Sauna, Calcagno, Ambudkar and Gottesman2007). For example, a transition from G to A at the fifth position of intron-32 of the dystrophin gene causes a splicing error leading to transcript termination (Tran et al., Reference Tran, Takeshima, Surono, Yagi, Wada and Matsuo2005).
Despite there being no evidence of variation in the amino acid sequence of K31, variation in the gene's promoter was found to be associated with a number of wool traits, including GFW, CFW and MSL. The presence of C in a sheep's genotype was associated with an average additional 200 g of CFW. This effect seemed to be consistent with the findings for MSL, suggesting the increase in GFW and CFW may have come about because of increased fibre growth, as opposed to an increase in fibre diameter traits. This effect was similar to the findings reported by Gong et al. (Reference Gong, Zhou, Hodge, Dyer and Hickford2015) with the KAP1-2 gene. This may, however, simply reflect the observation made by Gong et al. (Reference Gong, Zhou, Hodge, Dyer and Hickford2015) that MSL, CFW and GFW are moderately positively correlated traits.
The possibility exists that the effects observed for KRT31 may be due to its linkage to other KRTs and KRTAPs on the same chromosome. Seventeen other hair KRTs and KRTAPs have been identified on sheep chromosome 11 near KRT31 (Oar_v4.1 reference assembly), including KRT32, KRT33a, KRT33b, KRT34 − KRT36, KRT38 − KRT40, KRTAP3-3, KRTAP3-2, KRTAP1-1, KRTAP1-2, KRTAP1-3, KRTAP1-4, KRTAP4-3 and KRTAP4-1. These KRTs and KRTAPs are clustered, potentially variable and expressed in the wool fibre. Their proximity to each other might mean that it would be difficult to isolate and illustrate the independent effects of individual KRTs and KRTAPs.
Clean fleece weight and MFD are two important traits in wool production and selection for either CFW or MFD may lead to a change in the other trait. However, in the current study, while there was an association with variation in GFW, CFW and MSL, there was no association observed with MFD. This suggests that selecting for improvements in GFW and CFW may be possible, without having an adverse effect on MFD. This is consistent with the conclusions of Gong et al. (Reference Gong, Zhou, Hodge, Dyer and Hickford2015), and supported by the Trangie QPLU$ project (CSIRO, Australia), which has revealed that both CFW and MFD can be improved concurrently using genetic selection (Mortimer et al., Reference Mortimer, Taylor, Atkins and Pope2006). It also would require further testing in more sheep of different breeds, genders and ages.
Financial support
The current work was in part supported by funding from the Foundation for Research Science and Technology (FRST) (C10X0710), Basic Research Creative Groups of Gansu Province (18JR3RA190) and the Lincoln University Gene-Maker Laboratory. We thank the support of the AGMARDT Postdoctoral Fellowship to Hua Gong, and the New Zealand Guardian Trust for the Vernon Willey Trust Fellowship to HZ.
Conflict of interest
None.
Ethical standards
Not applicable.