Introduction
The WW domain is composed of 38–40 semi-conserved amino acid residues that mediate protein–protein interactions and are functionally similar to the SH3 domain (Chen et al., Reference Chen, Einbond, Kwak, Linn, Koepf, Peterson, Kelly and Sudol1997; Meiyappan et al., Reference Meiyappan, Birrane and Ladias2007), a model of protein–protein interactions. WW domain binding protein 1-like (WBP1L), a transmembrane adaptor protein (Guan et al., Reference Guan, Ni, Han, Lin, Zhang, Chen, Zhu, Liu and Zhang2020), can act as a regulator to identify a prognostic factor known as acute leukaemia 1 (OPAL 1) (Mosquera-Caro et al., Reference Mosquera-Caro, Helman, Veroff, Shuster, Martin, Davidson, Potter, Harvey, Hromas, Andries, Atlas, Wilson, Ar, Xu, Chen, Carroll, Camitta and Willman2003; Neveu et al., Reference Neveu, Spinella, Richer, Lagacé, Cassart, Lajoie, Jananji, Drouin, Healy, Hickson and Sinnett2016), binding to a variety of receptors in the body to regulate the expression level of related materials. Sequence analysis showed that WBP1L is a transmembrane aptamer protein. It has short extracellular and luminal region, and can form a small compact domain with a single transmembrane domain and a large cytoplasmic tail (Pei and Grishin, Reference Pei and Grishin2012). It has been found that mice deficient in WBP1L regulate B cell development and improve the efficiency of bone marrow cell transplantation (Borna et al., Reference Borna, Drobek, Kralova, Glatzova, Splichalova, Fabisik, Pokorna, Skopcova, Angelisova, Kanderova, Starkova, Stanek, Matveichuk, Pavliuchenko, Kwiatkowska, Protty, Tomlinson, Alberich-Jorda, Korinek and Brdicka2020). In addition, smoking causes WBP1L methylation in a pregnant woman's placenta, which regulates birth weight of a baby (Cardenas et al., Reference Cardenas, Lutz, Everson, Perron, Bouchard and Hivert2019). Therefore, we hypothesized that this gene might affect cattle growth traits. Most of the research on WBP1L gene is focused on humans. However, the detailed function of WBP1L gene in livestock and poultry has not been reported.
China has a vast territory, complex climate and terrain, a wide variety of organisms, and extremely rich genetic resources. However, the shortcomings of local varieties are particularly obvious, which are highlighted in slow growth rate, low feed utilization rate, low lean meat rate and high fat content. Therefore, in order to meet people's higher consumption demand and improve the economic benefits of breeding industry, we must breed superior individuals using some technical method and improve their growth performance.
As a kind of gene mutation, copy number variations (CNVs) have been widely used in the genetic improvement of livestock and poultry (Cheng et al., Reference Cheng, Jiang, Cao, Huang, Lan, Lei, Hu and Chen2020). CNVs are chromosomal structural variations that are greater than 50 bp in length in the genome (Iafrate et al., Reference Iafrate, Feuk, Rivera, Listewnik, Donahoe, Qi, Scherer and Lee2004), including duplicates, deletions, transformations, inversions and large-scale insertions, of which the most common type is duplicates (Feuk et al., Reference Feuk, Carson and Scherer2006; MacDonald et al., Reference MacDonald, Ziman, Yuen, Feuk and Scherer2014). CNVs are characterized by high resolution, large genome coverage and stable genetic performance. In recent years, a large number of gene CNVs have been found to be related to the growth characteristics of livestock and poultry, which has great significance for livestock breeding and genetic breeding. However, the relationship between CNVs of WBP1L gene and beef cattle growth traits has not been studied so far.
The WBP1L gene is located on chromosome 26 of cattle and has a total length of 69 985 bp. The known CNV was detected by quantitative polymerase chain reaction (qPCR) and its copy number was determined by the $2 \times 2^{-{\rm \Delta \Delta }C_{\rm t}}$![]() method. In the current study, CNV of the WBP1L gene was correlated with growth traits in 732 cattle from seven breeds (Qinchuan cattle, QC; Pinan cattle, PN; Yuengling cattle, YL; Xianan cattle, XN; Jiaxian cattle, JX; natural Guyuan cattle, NGY; Jian cattle, JA). However, the main subjects in this study were PN, JX, NGY and JA cattle, while the other three breeds were used as control experiments for data comparison. The results showed that CNV of WBP1L gene could be used as an important marker for PN, JX, NGY and JA molecular breeding.
method. In the current study, CNV of the WBP1L gene was correlated with growth traits in 732 cattle from seven breeds (Qinchuan cattle, QC; Pinan cattle, PN; Yuengling cattle, YL; Xianan cattle, XN; Jiaxian cattle, JX; natural Guyuan cattle, NGY; Jian cattle, JA). However, the main subjects in this study were PN, JX, NGY and JA cattle, while the other three breeds were used as control experiments for data comparison. The results showed that CNV of WBP1L gene could be used as an important marker for PN, JX, NGY and JA molecular breeding.
Materials and methods
Cattle and growth trait measurements
Previous sequencing revealed that CNV of WBP1L gene existed in cattle population. In this study, 732 individuals from seven Chinese cattle breeds were selected to analyse the correlation between growth characteristics of cattle breeds and CNVs of WBP1L gene. They are QC (n = 85), PN (n = 129), YL (n = 101), XN (n = 121), JX (n = 122), NGY (n = 113) and JA (n = 61). QC are one of the five big cattle breeds in China, large in size, strong in service and good in producing meat. It is named for the eight-hund-li Qin Chuan, which is produced in Guanzhong area of Shaanxi province. PN is a breed of hybrid cattle between Piedmont cattle and Nanyang cattle. It has delicious meat and high nutritional value. YL is the first breed of Chinese cattle bred by three-way crossbreeding in China, which has wide adaptability and excellent performance. XN is a new breed of Chinese cattle with Charolai cattle as its male parent and Nanyang cattle as its female parent. It is also the first Chinese cattle breed with independent intellectual property rights in China. JX originated in Jiaxian county, Henan province, and is a famous local cattle breed with both service and meat. NGY, a national geographical indication product of China, is produced in Guyuan city, Ningxia Hui autonomous region. JA is one of the three local fine breed cattle in Jiangxi province, which has good endurance and resistance to adversity.
All animal experiments complied with the ARRIVE guidelines and were carried out in accordance with the UK Animals (Scientific Procedures) Act, 1986 and associated guidelines, EU Directive 2010/63/EU for animal experiments, or the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). In addition, the Animal Experiment Review Committee of Northwest A&F University approved all of our experiments. Based on the International Animal Guidelines, we strictly adhere to animal testing policies and safeguard animal welfare. During the same breeding period, our experimental cattle were fed with the same nutritional conditions and the same management methods of feeding and drinking water. The correlation between CNV of WBP1L gene and the growth traits of 732 cattle of seven breeds was studied in a randomized experimental population. It should be noted that none of the cattle in the study were related.
Sampling and DNA extraction of cattle tissue
We put the extracted ear sample into the sample tube filled with alcohol and shipped it immediately back to the laboratory. During the extraction of genomic DNA, the ear samples collected were first homogenized and fully lysed with cell lysate, then the proteins were removed by protease, chloroform was extracted once, preparation solution was precipitated, RNA was removed by RNase and ethanol was precipitated to obtain the complete DNA. We diluted the DNA to 25 ng/μl and stored in a refrigerator at −40°C over a long period of time. Prior to the experiment, genomic DNA integrity was determined by agarose gel electrophoresis and its concentration was determined by a Nanodroplet 2000 ultramicrophotometer (Thermoscience, Waltham, MA, USA).
Primer design and amplification detection
After genome sequencing and analysis of the CNV area of WBP1L gene, the cattle WBP1L gene sequence (AC_000183.1) issued in NCBI was used as the reference sequence in this study, and the CNVR location between 23 641 732 and 23 643 331 bp was determined, with a total length of 1600 bp (Huang et al., Reference Huang, Li, Wang, Yu, Cai, Zheng, Li, Zhang, Chen, Asadollahpour Nanaei, Hanif, Chen, Fu, Li, Cao, Zhou, Liu, He, Li, Chen, Chen, Lei, Liu and Jiang2021) (Fig. 1). Primers were designed in CNVR according to the restriction conditions, and multiple pairs of primers were designed for PCR experiment. Primer information is shown in Table 1.

Fig. 1. (Colour online) Region of WBP1L gene CNV in cattle. Blue area stands for the coding area. The CNV area is from 23 641 732 to 23 643 331 in Chr26. Primer matching begins from 23 371 659 to 23 371 803 and the detected target sequence length of CNV is 145 bp. In addition, the total length of the CNV region is 1600 bp.
Table 1. Primers in correlation experiments

Agarose gel electrophoresis was used to detect their specificity; next the best pair of primers was selected. We selected BTF3 gene as our experiment internal reference gene. Using the same method, our primers were designed. The reaction system was as follows: 5 μl 2× Taq PCR Master Mix, 0.4 μl for each primer, 3 μl ddH2O and 1 μl DNA sample to form a 10 μl premixture system. PCR procedures for: 95°C 2 min, 95°C 10 s, 62°C 10 s, 72°C 10 s, 72°C for 10 min, 10°C up, extending stage circulation 35 times, and then use 1.5% agarose gel electrophoresis to detect primers for the specificity of the amplification products.
Experimental process and method
We repeated individual quantitative experiments three times for each animal, and then sent to company to obtain measurement data, viewing the data provided by CFX software; the same method was used to perform a simple comparison between individual's three sets of data, by removing two other significant difference of the data, and then calculated the average. The body size data of individual animals were matched with the analysis results. We used Excel software combined with $2 \times 2^{-{\rm \Delta \Delta }C_{\rm t}}$![]() to probe into the copy number of WBP1L gene in bovine genome. The correlation between CNV of WBP1L gene and growth traits of each animal was analysed using SPSSV23.0 software (SPSS, Inc., Chicago, IL, USA) with CNV type as the fixed factor and body size traits as the dependent variable.
to probe into the copy number of WBP1L gene in bovine genome. The correlation between CNV of WBP1L gene and growth traits of each animal was analysed using SPSSV23.0 software (SPSS, Inc., Chicago, IL, USA) with CNV type as the fixed factor and body size traits as the dependent variable.
Statistical analysis
This study tests the CNV of WBP1L gene and the correlation between bovine growth characters, and uses the $2 \times 2^{-{\rm \Delta \Delta }C_{\rm t}}$![]() method to calculate WBP1L gene in the bovine genome copy number; the final result for ‘0’ or ‘1’ was represented with the letter ‘Deletion’, the results for ‘2’ with the letter ‘Normal’, the results of larger than 2 were expressed with the letter ‘Duplication’ (for more than 20 sets of data, no follow-up analysis was performed). In order to analyse the influence of CNV of WBP1L gene on the phenotypic value of the trait, and to ensure the minimum effect of various factors on this experiment, we established the following simplified mathematical model:
method to calculate WBP1L gene in the bovine genome copy number; the final result for ‘0’ or ‘1’ was represented with the letter ‘Deletion’, the results for ‘2’ with the letter ‘Normal’, the results of larger than 2 were expressed with the letter ‘Duplication’ (for more than 20 sets of data, no follow-up analysis was performed). In order to analyse the influence of CNV of WBP1L gene on the phenotypic value of the trait, and to ensure the minimum effect of various factors on this experiment, we established the following simplified mathematical model:
where Zijk is the phenotype value of traits of experimental cattle; μ is the average value of each experimental cattle trait, CNVi is the effect of copy number variation type of WBP1L gene, Xi is the influencing factor of gender, Yi is the influencing factor of age and eijk is the random residual error. Furthermore, the experimental results were expressed as mean ± s.e. values.
Results
Expression of WBP1L gene in XN cattle
To detect the expression level of WBP1L gene in Chinese cattle, XN cattle breed was selected for expression profile analysis (Fig. 2). The research data show that the expression level of WBP1L gene was highest in the kidney, followed by the liver, intestines and lung, which had similar and low expression levels.

Fig. 2. (Colour online) Expression profiling of WBP1L in XN cattle.
Distribution of CNV types in seven Chinese cattle breeds
Previous studies have confirmed that the gene CNV and livestock growth traits are correlated; in order to study this correlation, the copy number distribution of seven cattle breeds was examined, including QC, PN, YL, XN, JX, NGY and JA. According to the $2 \times 2^{-{\rm \Delta \Delta }C_{\rm t}}$![]() values, the types of CNV were classified into deletion type, normal type and duplication type. As shown in Fig. 3(a), the CNV of WBP1L gene of seven cattle breeds is concentrated in less than 0. As shown in Fig. 3(b), the CNV of WBP1L gene was more between 0 and 4 copies and less between 5 and 20 copies. The results showed that the deletion type individuals of PN (77%), NGY (81%) and JA (49%) were significantly higher than the other two types. Surprisingly, YL without deletion and normal type individuals, duplication type individuals were up to 100%, NGY without duplication type individuals, deletion type individuals were up to 81%.
values, the types of CNV were classified into deletion type, normal type and duplication type. As shown in Fig. 3(a), the CNV of WBP1L gene of seven cattle breeds is concentrated in less than 0. As shown in Fig. 3(b), the CNV of WBP1L gene was more between 0 and 4 copies and less between 5 and 20 copies. The results showed that the deletion type individuals of PN (77%), NGY (81%) and JA (49%) were significantly higher than the other two types. Surprisingly, YL without deletion and normal type individuals, duplication type individuals were up to 100%, NGY without duplication type individuals, deletion type individuals were up to 81%.

Fig. 3. (Colour online) Distribution of WBP1L gene copy variation in seven cattle breeds (a). The percentage of WBP1L gene CNV copy variation in seven cattle breeds (b).
Correlation analysis between CNV and growth traits of experimental cattle
With the development of molecular breeding technology, CNV molecular markers have gradually attracted people's attention. According to previous studies, CNV is closely related to the growth traits of livestock and poultry. In this study, SPSSV23.0 software (SPSS, Inc., Chicago, IL, USA) was used to analyse the correlation between different copy number types and different cattle growth traits. The results showed that the copy number of WBP1L was significantly correlated with heart girth (HG) of PN cattle (**P < 0.01), and the growth traits of deletion type cattle were significantly better than those of normal type and duplication type cattle (Table 2). The withers height (WH), body length (BL), rump length (RL), chest depth (CD) and body weight (BW) of JX cattle were significantly correlated with the copy number types (*P < 0.05), and the growth traits of normal type and duplication type cattle were significantly better than deletion type ones, but there was no significant difference in the growth traits between normal type and duplication type cattle (Table 3). WH of NGY cattle were significantly correlated with copy number types (*P < 0.05), and the growth traits of deletion type individuals were the worst (Table 4). WH of JA cattle were significantly correlated with copy number types (*P < 0.05), and the growth traits of normal type individuals were the best, while those of duplication type individuals were the worst (Table 5). Surprisingly, no significant correlation was found between the copy number types of QC, YL and XN and growth traits in this study.
Table 2. Statistical correlation analysis of growth traits in PN cattle with WBP1L-CNV

HAHC, height at hip cross; HW, hip width.
Note: The BW is the estimated value, and the estimation method is the square of HG × BL/10 800.
Table 3. Statistical correlation analysis of growth traits in JX cattle with WBP1L-CNV

HLW, hucklebone width; CW, chest width.
Table 4. Statistical correlation analysis of growth traits in NGY cattle with WBP1L-CNV
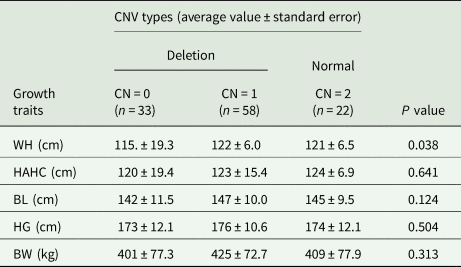
Note: The BW is the estimated value, and the estimation method is the square of HG × BL/10 800.
Table 5. Statistical correlation analysis of growth traits in JA cattle with WBP1L-CNV

CBC, cannon bone circumference.
Note: The BW is the estimated value, and the estimation method is the square of HG × BL/10 800.
Discussion
At present, people's understanding of WBP1L gene is not deep enough, and the research field focuses on the study of human diseases. For example, Andreassen et al. found that WBP1L gene is related to human schizophrenia (Andreassen et al., Reference Andreassen, Djurovic, Thompson, Schork, Kendler, O'Donovan, Rujescu, Werge, van de Bunt, Morris, McCarthy, Roddey, McEvoy, Desikan and Dale2013). Zhang et al. found a certain association between WBP1L gene and human hypertension (Zhang et al., Reference Zhang, Mo, Zhou, Zhu, HuangFu, Guo and Zhang2019). However, no related studies have been found, and we hope to discover the potential role of the gene in animal studies.
CNV is caused by genome rearrangement, which refers to the increase or decrease of the copy number of large segments (Zhang et al., Reference Zhang, Jiang and Gao2011; Vissers and Stankiewicz, Reference Vissers and Stankiewicz2012). It is mainly manifested as deletion or duplication at the submicroscopic level. These mutations are not only individual normal polymorphisms, but also pathogenic mutations, which are widely distributed in livestock and poultry genomes (Gheyas et al., Reference Gheyas and Burt2013; Zhao et al., Reference Zhao, Dong, Jiang, Wang, Xie, Rashid, Liu, Li, Bu, Wang, Ma, Sun, Wang, Bo, Zhou and Kong2020). Single-nucleotide polymorphisms (SNPs) and CNVs are two important potential sources of phenotypic variation in livestock and poultry, and the mutation rate of CNV sites is higher than that of SNPs. It is one of the most common types of variation. However, it should be pointed out that there are still many problems in the study of CNV: (1) due to the limitation of detection technology level, the detection effectiveness of small CNVs is limited; (2) due to the immaturity of relevant technologies and the lack of research data, the discovery rate of CNVs is much lower than SNP at present and (3) the CNV location reported at present is the location of a region containing CNV, and there are many possible mutations. There is still no good way to measure the exact mutation location or gene. All these will affect the research progress of CNV.
While studying the correlation between CNVs and phenotypes, some studies indicated that SNP genotypes and CNV measurements were statistically significantly correlated with 83 and 18% of gene-expression traits, respectively, but this underestimated the role of CNVs. Therefore, more complete and accurate detection techniques need to be proposed. Although CNV research is still incomplete, it has made many achievements: in human diseases (Henrichsen et al., Reference Henrichsen, Chaignat and Reymond2009), CNV has been used to study multiple myeloma (Yang et al., Reference Yang, Lv, Zhang, Li, Zhou, Lan, Lei and Chen2017a, Reference Yang, Wang, Zhu, Chang, Li, Chen, Long, Zhang, Liu, Lu and Qin2017b); leprosy (Ali et al., Reference Ali, Srivastava, Chopra, Aggarwal, Garg, Bhattacharya and Bamezai2013) and autism (Glessner et al., Reference Glessner, Wang, Cai, Korvatska, Kim, Wood, Zhang, Estes, Brune, Bradfield, Imielinski, Frackelton, Reichert, Crawford, Munson, Sleiman, Chiavacci, Annaiah, Thomas, Hou, Glaberson, Flory, Otieno, Garris, Soorya, Klei, Piven, Meyer, Anagnostou, Sakurai, Game, Rudd, Zurawiecki, McDougle, Davis, Miller, Posey, Michaels, Kolevzon, Silverman, Bernier, Levy, Schultz, Dawson, Owley, McMahon, Wassink, Sweeney, Nurnberger, Coon, Sutcliffe, Minshew, Grant, Bucan, Cook, Buxbaum, Devlin, Schellenberg and Hakonarson2009). In animal breeding, such as IGF1R gene of CNV and Jin Na salience (Jacqueline Nottingham) cow's weight and the height of QC and bone was significantly associated with wide (Ma et al., Reference Ma, Wen, Cao, Cheng, Huang, Ma, Hu, Lei, Qi, Cao and Chen2019), the CADM2 gene CNV of Guizhou black goats was obviously correlated with its shoulder height and BL, which lack individual in WH and BL of replication and normal individuals (Xu et al., Reference Xu, Wang, Zhang, An, Wen, Wang, Liu, Li, Lyu, Li, Wang, Ru, Xu and Huang2020), the CNV of MLLT 10 gene had significant effects on the HW and RL of cattle (*P < 0.05) (Yang et al., Reference Yang, Zhang, Xu, Qu, Lyv, Wang, Cai, Li, Wang, Xie, Ru, Xu, Lei, Chen, Huang and Huang2020). By inserting a 19 bp repeat sequence into the promoter region of CDKN3 gene, it was found that the growth and carcase traits of chicken were significantly affected (Li et al., Reference Li, Liu, Tang, Li, Han, Tian, Li, Li, Li, Liu, Kang and Li2019). The insertion mutation of lncFAM 97 bp was closely related to the growth traits of chickens at different growth stages (Li et al., Reference Li, Jing, Cheng, Wang, Li, Han, Li, Li, Sun, Tian, Liu, Kang and Li2020). In addition, CNV has been used for breeding in sheep (Jenkins et al., Reference Jenkins, Goddard, Black, Brauning, Auvray, Dodds, Kijas, Cockett and McEwan2016; Yang et al., Reference Yang, Xu, Zhou, Liu, Wang, Kijas, Zhang, Li and Liu2018) and monkeys (Boyd et al., Reference Boyd, Boyd and Brown1980). Our study found that CNV of WBP1L gene was significantly correlated with growth traits of a variety of Chinese yellow cattle.
In order to detect the relationship between different copy numbers of WBP1L gene and growth performance of QC, a total of 732 Chinese cattle breeds were tested. Of all the experimental results, there are four varieties showed significant correlation, and only QC HG a character showed significant correlation with the gene copy number, four varieties of nine characters showed significant correlation, but only a HG of Jiaxian red cattle breed attained up to five traits that showed significant correlation, WH was significantly correlated with JX, NGY and JA, and BL and BW were also significantly correlated with PN and JX populations. In addition, different from other populations, BL and CD were correlated in JX population. Among the seven herds, some had the best CNV of the normal type, some had the best CNV, while others had the best deletion type.
Conclusions
In conclusion, our study found that the CNV of WBP1L gene was obviously correlated with ten traits of four breeds of cattle, which in a sense indicated that these traits could be used for cattle seed selection, promoting breed improvement and promoting genetic and breeding work.
Author contributions
Methodology: J. Z. and Y. H.; software: Z. Z.; validation: X. L. and Y. C.; formal analysis: P. Y. and J. L.; investigation: Y. H. and L. L.; resources and data curation: W. H., G. Y. and Z. Q.; writing – original draft preparation: J. Z. and Y. H.; writing – review and editing: F. C., Z. L. and C. L.: visualization and supervision: Q. S. and B. R.; project administration and funding acquisition: E. W. and Y. H.
Financial support
This research has been supported by the Henan Beef Cattle Industrial Technology System (S2013-08); Science-Technology Foundation for innovation and creativity of Henan Academy of Agricultural Sciences (2020CX09); National Beef Cattle and Yak Industrial Technology System (CARS-37); The National Key Research and Development Program of China (2018YFD0501700).
Conflict of interest
The authors declare no conflict of interest.
Ethical standards
All animal experiments complied with the ARRIVE guidelines and were carried out in accordance with the UK Animals (Scientific Procedures) Act, 1986 and associated guidelines, EU Directive 2010/63/EU for animal experiments, or the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). In addition, the Animal Experiment Review Committee of Northwest A&F University approved all of our experiments. Based on the International Animal Guidelines, we strictly adhere to animal testing policies and safeguard animal welfare.











