INTRODUCTION
Colour is very important for the alpaca fibre industry because it has a substantial impact on the value of the fibre. Alpacas have more than 22 recognized colour variants and this fact, combined with the paucity of information on alpaca fibre colour inheritance patterns, and subjective phenotype recording methods, have resulted in inefficient prediction of colour outcomes in breeding programmes (Sponenberg Reference Sponenberg2001; McGregor Reference McGregor2006).
Pigmentation is a complex process with the potential for colour to be affected at any stage from the specification of melanocyte lineage at the neural crest to the export of melanin from the melanocyte (Hoekstra Reference Hoekstra2006; Thiruvenkadan et al. Reference Thiruvenkadan, Kandasamy and Panneerselvam2008). Although there are over 300 identified genes that have a known role in mammalian pigmentation, a few key genes have been identified as major regulators of pigment production in mammals (Rieder et al. Reference Rieder, Taourit, Mariat, Langlois and Guerin2001; Fontanesi et al. Reference Fontanesi, Beretti, Riggio, Dall'Olio, Calascibetta, Russo and Portolano2010). These are the melanocortin-1 receptor (MC1R), the α-melanocyte stimulating hormone (α -MSH) and the Agouti signalling protein (ASIP) genes (Sturm et al. Reference Sturm, Teasdale and Box2001; Rouzaud & Hearing Reference Rouzaud and Hearing2005; Hoekstra Reference Hoekstra2006). These three genes have been characterized extensively in many mammalian species including mice, humans, horses, sheep, cows, pigs and dogs, and many mutations have been reported that have associations with phenotype variation (Jackson et al. Reference Jackson, Budd, Horn, Johnson, Raymond and Steel1994; Rouzaud et al. Reference Rouzaud, Martin, Gallet, Delourme, Goulemot-Leger, Amigues, Menissier, Leveziel, Julien and Oulmouden2000; Rieder et al. Reference Rieder, Taourit, Mariat, Langlois and Guerin2001; Sturm et al. Reference Sturm, Teasdale and Box2001; Kerns et al. Reference Kerns, Olivier, Lust and Barsh2003).
Mammals are only able to produce two types of pigment, eumelanin and pheomelanin, which results in black to brown and red to yellow colours, respectively (Furumura et al. Reference Furumura, Sakai, Abdel-Malek, Barsh and Hearing1996; Rees Reference Rees2003; Hoekstra Reference Hoekstra2006). The protein products of the MC1R, α -MSH and ASIP genes, and the interactions between them, control the relative amount, type and location of pigment that is produced (Furumura et al. Reference Furumura, Sakai, Abdel-Malek, Barsh and Hearing1996; Rieder et al. Reference Rieder, Taourit, Mariat, Langlois and Guerin2001; Rees Reference Rees2003). Therefore, MC1R, α -MSH and ASIP are the genes primarily responsible for the wide array of pigment variation observed in mammals (Sturm et al. Reference Sturm, Teasdale and Box2001; Hoekstra et al. Reference Hoekstra, Hirschmann, Bundey, Insel and Crossland2006).
The product of the MC1R gene is the melanocortin-1 receptor protein (MC1R), which is expressed on the cell surface of melanocytes (Scott et al. Reference Scott, Wakamatsu, Ito, Kadekaro, Kobayashi, Groden, Kavanagh, Takakuwa, Virador, Hearing and Abdel-Malek2002; Newton et al. Reference Newton, Smit, Barnes, Pedley, Parsons and Sturm2005). Initiation of intracellular MC1R signalling is induced by the binding of its ligand, the protein product of α -MSH, α-MSH (Hearing Reference Hearing2005; Hoekstra Reference Hoekstra2006; Tully Reference Tully2007). The MC1R/α-MSH complex activates the adenyl cyclase pathway that leads to increased cyclic-adenosine mono-phosphate (cAMP) levels within the cell. A high level of cAMP is a crucial factor in the initiation of downstream signalling events within the cell leading to stimulation of eumelanin production (Buscà & Ballotti Reference Buscà and Ballotti2000; Oyehaug et al. Reference Oyehaug, Plahte, Vage and Omholt2002).
The ASIP gene encodes for a small, secreted protein, ASIP, c. 133 amino acids in length which consists of a secretion signal, a lysine-rich basic region and a folded, cysteine rich C-terminus (Hustad et al. Reference Hustad, Perry, Siracusa, Rasberry, Cobb, Cattanach, Kovatch, Copeland and Jenkins1995; Willard et al. Reference Willard, Bodnar, Harris, Kiefer, Nichols, Blanchard, Hoffman, Moyer, Burkhart, Weiel, Luther, Wilkinson and Rocque1995). ASIP is an antagonist of MC1R, and acts specifically to block the activity of the MC1R agonist, α-MSH, and inhibit MC1R activation (Hustad et al. Reference Hustad, Perry, Siracusa, Rasberry, Cobb, Cattanach, Kovatch, Copeland and Jenkins1995; Willard et al. Reference Willard, Bodnar, Harris, Kiefer, Nichols, Blanchard, Hoffman, Moyer, Burkhart, Weiel, Luther, Wilkinson and Rocque1995). When the receptor is in its inactive conformation, cAMP production is reduced or inhibited, and the cell switches to pheomelanin synthesis (Furumura et al. Reference Furumura, Sakai, Abdel-Malek, Barsh and Hearing1996; Le Pape et al. Reference Le Pape, Wakamatsu, Ito, Wolber and Hearing2008).
The ASIP gene consists of three coding exons, most often termed exons 2, 3 and 4. Many mammals have no reported mutations within the coding region, and in most species the coding region is highly conserved (Siracusa Reference Siracusa1994; Chen et al. Reference Chen, Duhl and Barsh1996; Graphodatskaya et al. Reference Graphodatskaya, Joerg and Stranzinger2002; McNulty et al. Reference McNulty, Jackson, Thompson, Chai, Gantz, Barsh, Dawson and Millhauser2005; Royo et al. Reference Royo, Alvarez, Arranz, Fernandez, Rodriguez, Perez-Pardal and Goyache2008; Fontanesi et al. Reference Fontanesi, Beretti, Riggio, Dall'Olio, Calascibetta, Russo and Portolano2010). However, non-agouti black in dogs and in Japanese quail is the result of mutation in exon 4 (Kerns et al. Reference Kerns, Newton, Berryere, Rubin, Cheng, Schmutz and Barsh2004; Hiragaki et al. Reference Hiragaki, Inoue-Murayama, Miwa, Fujiwara, Mizutani, Minvielle and Ito2008).
Recent research into alpaca fibre colour genetics has included studies on MC1R (Powell et al. Reference Powell, Moss, Tree, Roeder, Carleton, Campbell and Kooyman2008; Feeley & Munyard Reference Feeley and Munyard2009). Several polymorphisms were identified in MC1R that may influence colour in alpacas; however, these polymorphisms were not sufficient to explain all coat colour variation observed in alpacas. There has been no reported research investigating the alpaca ASIP gene and its effects on fibre pigment production. Due to its close interaction with MC1R, information about ASIP is essential for gaining an understanding of the genetic mechanisms controlling colour inheritance in alpacas, an idea that is supported by the fact that alpacas exhibit phenotypes analogous to phenotypes controlled by ASIP in other species (for example black and bay in horses; black and black and tan in dogs). The objectives of the current research were to analyse the coding region of the alpaca ASIP gene for mutations and ascertain their potential effect on pigment production.
MATERIALS AND METHODS
Animals and DNA extraction
Blood samples were collected from 94 alpacas (Table 1). Initial sequence analysis was carried out on 15 animals (three black, two black and tan, five brown and five fawn). An additional 79 animals, comprising a wider range of colour phenotypes, were subsequently analysed, but only for exon 4 mutations. Fibre colour was determined according to the owners’ assessment of the animal. Samples were collected from animals bred in the states of Western Australia, New South Wales and Victoria in Australia. Genomic DNA was extracted from 200 μl of EDTA anti-coagulated blood using the AxyPrep Blood Genomic DNA Miniprep Kit (Axygen, Union City, CA, USA) according to the manufacturer's instructions.
Table 1. Colour phenotypes of alpacas used in the current study

Amplification and sequencing of alpaca ASIP and MC1R
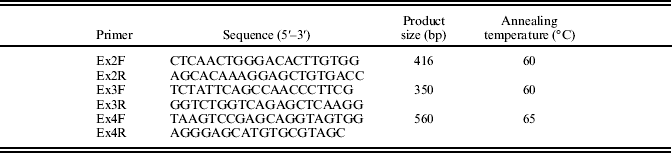
Polymerase chain reaction (PCR) primers were designed to amplify the three coding exons of the alpaca ASIP gene (Table 2). These primers were based on the alpaca sequence assembly available on the Ensembl database (http://www.ensembl.org/index.html) and were designed to hybridize c. 100 bp outside of the predicted splice sites for these exons, thereby amplifying the complete coding region of alpaca ASIP, and part of the introns of the gene. All PCRs were carried out in an Eppendorf Mastercycler (Eppendorf, North Ryde, New South Wales, Australia), in 10 μl reactions containing 67 mmol/l Tris/HCl (pH 8·8), 16·6 mmol/l [NH4]2SO4, 0·45% (v/v) Triton X-100, 0·2 mg/ml gelatin, 0·2 mmol/l dNTP (Fisher Biotec, Wembley, Western Australia, Australia), 0·2 μmol/l each of forward and reverse primer, 1 unit Taq DNA polymerase (Fisher Biotec), 1·5 mmol/l MgCl2 and 20 ng genomic DNA. Thermal cycles were: initial denaturation at 95°C for 3 min, followed by 30 cycles, each consisting of 94°C for 30 s, annealing for 30 s (Table 2) and 72 °C for 45 s; with a final extension at 72°C for 10 min. Amplified DNA was electrophoresed in 1·5% (w/v) agarose gels in TAE buffer, stained with ethidium bromide and visualized by UV transillumination. The PCR products were purified using the AxyPrep PCR Cleanup Kit (Axygen). Amplification of ASIP coding exons from each animal was carried out in five independent 10 μl reactions, which were pooled before purification and sequencing. Sequencing reactions were carried out using ASIP primers for each exon (Table 2) with Big Dye Terminator Technology v3.1 (Applied Biosystems, Mulgrave, Victoria, Australia) and analysed on a 3730 DNA analyser (Applied Biosystems). Genotypes at MC1R were obtained (following the procedures outlined in Feeley & Munyard Reference Feeley and Munyard2009) for non-black alpacas in this study who exhibited putative non-functional ASIP mutations.
Table 2. Primer pairs designed for amplification of ASIP exons from genomic DNA
Sequence assembly and analysis
Splice sites were determined using the program SpliceView (http://zeus2.itb.cnr.it/~webgene/wwwspliceview.html; verified 19 Nov 2010) coupled with the known bovine and human ASIP exons. Complete ASIP coding region sequences for each animal were compiled using Geneious software (Biomatters, Auckland, New Zealand), and were compared with genes and proteins from other species by GenBank NCBI BLASTn and BLASTx protocols (http://blast.ncbi.nlm.nih.gov/Blast.cgi; verified 19 Nov 2010).
Genotyping ASIP mutations
Once mutations were identified in alpaca ASIP (Table 3) an additional sample group was genotyped for the four exon 4 polymorphisms (Table 4). The PCRs were carried out as above on an additional 79 animals (Table 1). Amplified DNA was electrophoresed in 1·5% (w/v) agarose gels in TAE buffer, stained with ethidium bromide and visualized by UV transillumination. PCR products were then sequenced using Big Dye v3.1 on a 3730 DNA analyser (Applied Biosystems) at Macrogen Inc., South Korea, using primers Ex4F and Ex4R. The deletion was analysed by electrophoresis in 2% (w/v) agarose gels in TAE buffer, stained with ethidium bromide and visualized by UV transillumination. Fisher's exact test was used to test for non-random association between mutations and colour.
Table 3. Polymorphisms identified in the alpaca ASIP gene
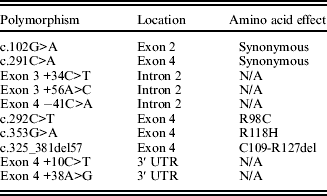
Table 4. ASIP genotypes of the three significant exon 4 polymorphisms examined in the current study

Predictive modelling of ASIP
A multiple alignment of ASIP sequences from 32 different mammalian species was performed using Jalview 2.0 (Waterhouse et al. Reference Waterhouse, Procter, Martin, Clamp and Barton2009) with the Muscle algorithm (Edgar Reference Edgar2004) using default values. The web server versions of the scale-invariant feature transform (SIFT; Kumar et al. Reference Kumar, Henikoff and Ng2009), iMutant 2.0 (Capriotti et al. Reference Capriotti, Fariselli and Casadio2005), iPTree-STAB (Huang et al. Reference Huang, Gromiha and Ho2007) and PolyPhen (Ramensky et al. Reference Ramensky, Bork and Sunyaev2002) algorithms were used to predict the potential effect of amino acid substitutions on ASIP structure and function. Default values were used for all programs. SIFT is based on sequence conservation and homology, using position-specific scoring matrices. It predicts the effects of amino acid substitution on both structure and function. iMutant 2.0 and iPTree-STAB use a thermodynamic method and base their predictions on protein stability using the calculated free energy change (ΔΔG) of mutations within the protein sequence. PolyPhen bases its predictions on empirical rules applied to the protein's sequence, phylogenetic and structural information. All methods, except iPTree-STAB, provide an estimate of confidence in their predictions. The signal peptide was detected using the web servers of SignalP (Bendtsen et al. Reference Bendtsen, Nielsen, von Heijne and Brunak2004), CoSiDe (http://sigpep.services.came.sbg.ac.at/; verified 19 Nov 2010) and Phobius (Kall et al. Reference Kall, Krogh and Sonnhammer2007).
RESULTS
The alpaca ASIP gene
The complete coding sequence of the alpaca ASIP gene was generated (GenBank accession no. HM768322). Homology with the ASIP gene of other species was confirmed, with the highest sequence homology being to cow, goat and sheep (89%) and pig (88%). Exon 2 is 160 bp, exon 3 is 65 bp and exon 4 is 177 nucleotides in length. The entire coding region is 402 bp long, 6 bp longer than the dog and mouse, 3 bp longer than the human and 6 bp shorter than the cat ASIP coding region. A consensus splice acceptor and splice donor site flanks each coding exon. Exons 2, 3 and 4 contain 54, 40 and 68% GC (respectively) and the whole coding region is 58% GC.
The predicted alpaca ASIP protein
Alpaca ASIP translates into a 133 amino acid predicted protein (Fig. 1) that is 0·83 identical to sheep and cow ASIP and 0·81 identical to the horse and rabbit proteins (see end of this paper for link to supplementary information). The consensus prediction for the signal peptide in ASIP was from residues 1 to 22 by two out of three signal peptide prediction methods. The predicted peptide cleavage site was between residues 22(S) and 23(H). In contrast, Phobius predicted a signal peptide cleavage site between residues 24 and 25.

Fig. 1. The alpaca ASIP gene. (a) Coding exons 2, 3 and 4 are shown as solid boxes. Untranslated regions are shown as dashed lines (not to scale). Polymorphisms are shown in the regions where they occur. Indicated intron size was determined from the Ensembl genome assembly. (b) The predicted alpaca wild-type ASIP protein. The conserved cysteine residues in the C-terminus are underlined, the secretion signal is shown as a dotted box. Predicted exon 4 protein sequences for the R98C, R118H (grey boxes) and C109_R127del mutations are shown below the wild-type protein.
A pairwise alignment of the alpaca ASIP sequence with the sequence of an engineered ASIP structure (Protein Data Base ID: 2KZA) showed that they were 0·74 identical in the 53 residue, cysteine rich, C-terminal sequence. All cysteines in the pairwise alignment were exactly aligned. Consequently, it can be inferred by sequence similarity that the disulphide bonds in alpaca ASIP are likely to occur at cysteine residue locations [94–109], [101–115], [108–126], [112–133] and [117–124]. The disulphide bond prediction method DBCP (Lin & Tseng Reference Lin and Tseng2010) also predicted disulphide bonds at exactly the same locations in alpaca ASIP.
Mutations in alpaca ASIP and MC1R
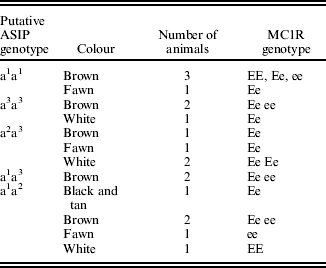
Sequencing of the ASIP coding region in 15 alpacas revealed five polymorphisms (Table 3). Two non-synonymous single nucleotide polymorphisms (SNPs) were identified in exon 4: c.C292T, predicted to cause an arginine-to-cysteine substitution at codon 98 (R98C; GenBank accession no. HM768323), and c.G353A, predicted to cause an arginine-to-histidine substitution at codon 118 (R118H; GenBank accession no. HQ008273). A 57 bp deletion was also discovered in exon 4. This deletion, occurring at nucleotides 325–381 (c.325_381del57; GenBank accession no. HM768324), is predicted to result in 19 of the last 25 amino acids being absent from the mature protein (p.C109_R127del). In addition, two synonymous mutations; c.G102A in exon 2 (p.G34) and c.C291A in exon 4 (p.T291) were identified. Four haplotypes involving these mutations were observed in this group of alpacas, and these haplotypes existed in ten different combinations (Table 4). While no match occurred between any fibre colour and any single ASIP genotype in the 94 animals studied (Table 4) there was strong association between the homozygous state of each mutation and black colour, as well as between the combined alleles and black colour (Fisher's exact test for c.325_381del57 P<0·001; for c.292C>T P=0·039; for c.353G>A P=0·024; and for all combined P<0·001). Forty eight of the 53 black animals were either homozygous for C109_R127del (allele a1), the T allele of R98C (a2), the A allele of R118H (a3) or were heterozygous for a combination of two of these mutations. However, 18 of the 41 non-black animals also exhibited some of these same genotype combinations. These animals exhibited a range of MC1R genotypes (Table 5). An additional five SNPs were identified in the non-coding regions flanking the exons (Table 3).
Table 5. MC1R Genotypes of non-black animals with aa genotypes
Predicted functional effect of ASIP mutations
The arginine residues at positions 98 and 118 in the alpaca ASIP sequence are highly conserved in all ASIP sequences aligned from 32 different mammalian species (see end of the paper for link to supplementary material). All amino acid substitution prediction methods used predicted that the R98C and R118H amino acid substitutions are potentially damaging to ASIP structure or function (Table 6). Only the SIFT method labelled its prediction as ‘low confidence’. Therefore, it is proposed that the mutations R98C, R118H and C109-R127del are all loss of function non-agouti equivalent ‘a’ mutations for alpaca ASIP.
Table 6. Potential effect of amino acid substitution on ASIP structure or function
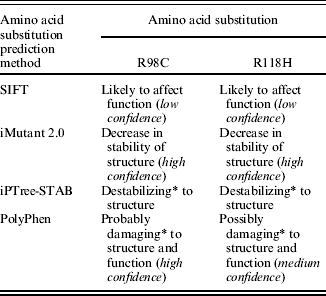
* The terms ‘destabilizing’, ‘probably damaging’ and ‘possibly damaging’ are used by the respective methods to describe the effect of the amino acid substitution.
DISCUSSION
Three novel mutations in exon 4 of the alpaca ASIP gene have been identified that are each predicted to cause a loss of function in the protein. Exon 4 codes for the last 40 amino acid residues of ASIP, which constitute the C-terminal domain and the majority of the residues are responsible for protein activity and receptor binding (Dinulescu & Cone Reference Dinulescu and Cone2000; Miltenberger et al. Reference Miltenberger, Wakamatsu, Ito, Woychik, Russell and Michaud2002). It has been reported that the ten cysteine residues within the C-terminus are involved in a network of five disulphide bonds that acts to stabilize the protein (McNulty et al. Reference McNulty, Jackson, Thompson, Chai, Gantz, Barsh, Dawson and Millhauser2005; Yu & Millhauser Reference Yu and Millhauser2007) and form a unique fold motif known as an inhibitor cysteine knot (ICK; McNulty et al. Reference McNulty, Jackson, Thompson, Chai, Gantz, Barsh, Dawson and Millhauser2005; Yu & Millhauser Reference Yu and Millhauser2007). The spacing of the ten cysteine residues in ASIP is strictly conserved throughout all mammals that have been investigated (Miltenberger et al. Reference Miltenberger, Wakamatsu, Ito, Woychik, Russell and Michaud2002; McNulty et al. Reference McNulty, Jackson, Thompson, Chai, Gantz, Barsh, Dawson and Millhauser2005; Yu & Millhauser Reference Yu and Millhauser2007). The particular fold structure of the ICK allows for presentation of three important conserved residues, Arg116Phe117Phe118, in order to facilitate MC1R interaction and binding (Miltenberger et al. Reference Miltenberger, Wakamatsu, Ito, Woychik, Russell and Michaud2002). This structure can therefore be assumed to be essential for correct ASIP functioning. Animals carrying C109_Rdel19 (aka a1) are missing six of these ten conserved cysteine residues. This would almost certainly prevent tertiary structures, such as the ICK, from being formed correctly. With this type of disruption to the protein it is almost certain that function is eliminated.
About half of all known disease-causing mutations result from amino acid substitutions, and automated prediction methods can be used to identify potentially damaging substitutions (Ng & Henikoff Reference Ng and Henikoff2006). The current study used four automated methods to derive a consensus view on the potential effect of the two observed amino acid substitutions on ASIP structure and function. It is preferable to use more than one prediction method, based on different assumptions and algorithms, because this gives increased support for any inference that may be made on the functional or structural significance of an amino acid substitution. In all cases the automated methods predicted a potentially damaging or destabilizing effect on ASIP structure or function for both SNPs (Table 6). Only the SIFT method labelled its prediction as ‘low confidence’. The SIFT method depends on sequence diversity to generate confidence values and there was insufficient sequence diversity in the sequence alignment generated by SIFT (which uses PsiBlast to find and align, functionally related sequences). This is a common issue with prediction algorithms that depend on sequence conservation and is understandable given the highly conserved nature of the ASIP sequence.
There are a number of ways in which these SNPs might cause a loss of function in ASIP. The R98C polymorphism results in the substitution of a cysteine for a histidine in the highly conserved C-terminus of the protein. Previous studies have reported that the loss of even one of these highly conserved cysteine residues is sufficient to abolish the activity of the protein (Perry et al. Reference Perry, Hustad, Swing, Jenkins and Copeland1995; Miltenberger et al. Reference Miltenberger, Wakamatsu, Ito, Woychik, Russell and Michaud2002). The addition of another cysteine residue in this region is predicted to disrupt the formation of this essential tertiary structure and be responsible for a loss of function of the protein. Protein alignment results suggest that the R98C polymorphism in alpacas is the same as the R96C polymorphism present in dogs (Kerns et al. Reference Kerns, Newton, Berryere, Rubin, Cheng, Schmutz and Barsh2004), due to the upstream difference of two amino acids between the two proteins. The R96C mutation in dogs has been shown to be a loss of function mutation resulting in non-agouti black (Kerns et al. Reference Kerns, Newton, Berryere, Rubin, Cheng, Schmutz and Barsh2004). The similarity between these polymorphisms further supports the current in silico predictions that R98C has a similar affect in alpacas.
The residues Arg116Phe117Phe118 have an essential role in protein interactions of the C-terminal domain (Miltenberger et al. Reference Miltenberger, Wakamatsu, Ito, Woychik, Russell and Michaud2002; McNulty et al. Reference McNulty, Jackson, Thompson, Chai, Gantz, Barsh, Dawson and Millhauser2005). When correctly folded, these are the binding determinants of the protein and they facilitate direct interaction with Melanocortin receptors (Miltenberger et al. Reference Miltenberger, Wakamatsu, Ito, Woychik, Russell and Michaud2002; McNulty et al. Reference McNulty, Jackson, Thompson, Chai, Gantz, Barsh, Dawson and Millhauser2005). This suggests that the amino acid sequence of this domain and the correct structural folds are important determinants for protein function. The R118H substitution is usually considered a conservative substitution according to the Blosum62 evolutionary matrix (Henikoff & Henikoff Reference Henikoff and Henikoff1992) and this is understandable given the similar physicochemical properties of arginine and histidine. However, any change in a highly conserved region has the potential to be detrimental to the protein's structure or function. The conserved Arg116Phe117Phe118 amino acids are homologous to residues Arg118Phe119Phe120 in alpaca ASIP. Therefore, the R118H mutation results in a His-Phe-Phe combination present in the active loop (instead of the conserved Arg-Phe-Phe combination) which may prevent ASIP interaction with MC1R, resulting in a loss-of-function variant. However, functional studies, such as testing the effect of these proteins on receptor signalling in vitro or RNAi in mouse models, are needed to confirm that these mutations are affecting ASIP function. It is interesting to note that the three exon 4 mutations probably occurred in historically independent populations. The R118H mutation cannot exist in combination with C109_T127del19, and the T allele of R98C was never found in combination with the deletion, nor with the A allele of R118H. Only four haplotypes were present in the sample of alpacas used in the current study: C,G,No, C,Yes, T,G,No and C,A,No. Black alpacas are considered to be sacred in South America, and are still sacrificed to the Gods (Bolin Reference Bolin1998; J. C. Wheeler, personal communication). Therefore, historically, any new mutations causing black animals to occur would have been preserved by selective breeding.
It would be expected that animals that are homozygous for any of the non-functional ASIP alleles would have a black phenotype. This is largely consistent with the current data (Table 4): 48 out of 53 black animals were homozygous for a1, a2 or a3, or were heterozygous for two of these alleles. The five animals that were black in phenotype but did not carry two putative black alleles could have as yet undiscovered mutations in ASIP exons 2 or 3, regulatory mutations leading to decreased ASIP expression, or dominant mutations at another gene (e.g. MC1R or β-defensin).
Almost half of the non-black animals were homozygous for the putative non-functional mutations. There are four reasons why non-black animals might carry a black genotype. The first is that a non-functional MC1R genotype is also present (i.e. ee), thus the specific ASIP allele is irrelevant. MC1R and ASIP have an epistatic relationship where a fully functioning MC1R receptor is necessary for the ASIP alleles to be expressed (Furumura et al. Reference Furumura, Sakai, Abdel-Malek, Barsh and Hearing1996; Hoekstra Reference Hoekstra2006). If MC1R is non-functional then it cannot be activated or inactivated by either of the two alternate ligands. It will therefore only express pheomelanin, and the ASIP alleles will be masked. Just over one-third of the non-black ‘aa’ animals are homozygous for the putative non-functional MC1R allele ‘e’ (Table 5; Feeley & Munyard Reference Feeley and Munyard2009); however, others were not. Secondly, if α-MSH is also non-functional, then MC1R will receive no signal from either its agonist or antagonist, and will revert to pheomelanin production.
A third possible explanation is that dilution genes are having an effect on the underlying colour. The dilution genes MATP and TYR have been investigated in alpacas, but no mutations were found that were linked with colour variation (Cransberg & Munyard Reference Cransberg, Munyard and Alex Safari2009). The mutation responsible for the Champagne dilution in horses has recently been identified in SLC36A1 (Cook et al. Reference Cook, Brooks, Bellone and Bailey2008). This mutation renders black animals brown, and chestnut animals pale golden. Another strong candidate dilution gene is TYRP1, which is known to change black eumelanin to brown eumelanin in many species (e.g. dogs: Schmutz et al. Reference Schmutz, Berryere and Goldfinch2002; cows: Berryere et al. Reference Berryere, Schmutz, Schimpf, Cowan and Potter2003; sheep: Gratten et al. Reference Gratten, Beraldi, Lowder, McRae, Visscher, Pemberton and Slate2007). The current authors have found no report of any investigation of these two genes in alpacas.
The fourth potential reason why ‘aa’ alpacas might be non-black is also a reason why some black animals are not ‘aa’: subjective phenotype assignment. In the current authors’ experience there is considerable variation in the words used to describe similarly coloured alpacas. For instance, one breeder would call a faded black animal brown, while another would call it black. Similar problems also occur when assigning colours to paler animals. It is imperative that the industry adopts an objective way to measure phenotype, perhaps following the simple melanin analysis methods developed by Ito et al. (Reference Ito, Wakamatsu and Ozeki2000).
While black alleles have been identified in the coding regions of the gene, there is almost certainly additional variation in the regulatory regions of alpaca ASIP that leads to other phenotypes in this species. Alpacas with phenotypes resembling black and tan dogs and bay horses are common, and samples of fibre banded in a hair-cycle specific manner have been observed. It has been shown in several studies that ASIP has a complex structure including different regulatory elements which are involved in expression of the gene in region-specific and cycle-specific ways (Dinulescu & Cone Reference Dinulescu and Cone2000; Fontanesi et al. Reference Fontanesi, Beretti, Riggio, Gomez Gonzalez, Dall'Olio, Davoli, Russo and Portolano2009). Royo et al. (Reference Royo, Alvarez, Arranz, Fernandez, Rodriguez, Perez-Pardal and Goyache2008) and Gratten et al. (Reference Gratten, Pilkington, Brown, Beraldi, Pemberton and Slate2010) demonstrated that the regulatory exons of ASIP are important in controlling expression in different breeds of sheep. Norris & Whan (Reference Norris and Whan2008) found that the white phenotype in sheep is dependent on a partial duplication of the regulatory region of ASIP.
The mutation found in exon 2 is a synonymous SNP resulting in no predicted structural changes, and it was polymorphic in every colour group represented in the current sample set. Therefore, although it occurs in the region that codes for the signal sequence and part of the lysine-rich basic region (Hustad et al. Reference Hustad, Perry, Siracusa, Rasberry, Cobb, Cattanach, Kovatch, Copeland and Jenkins1995; Willard et al. Reference Willard, Bodnar, Harris, Kiefer, Nichols, Blanchard, Hoffman, Moyer, Burkhart, Weiel, Luther, Wilkinson and Rocque1995), it is almost certainly only a population polymorphism. In conclusion, the three novel loss-of-function ASIP mutations identified in the current study can be used by alpaca breeders to decrease the frequency of black animals in their herd, by selecting against animals carrying any of the mutations. It will also allow breeders of black alpacas to retain the desired colour, increase their gene pool, and potentially improve the quality of fibre, by identifying high-quality non-black animals that carry black. However, these mutations neither explain all instances of black nor all the phenotypic variation that is possible in ASIP.
Supplementary material is available for this paper. http://journals.cambridge.org/AGS









