The definition of life problem
‘I am not going to answer this question. In fact, I doubt if it will ever be possible to have a full answer, because we know what it feels like to be alive, just as we know what redness, or pain, or effort are. So we cannot describe them in terms of anything else. But it is not a foolish question to ask.’ (Haldane Reference Haldane1949)
We instinctively believe that the words ‘Life’ and ‘Living’ relate to real and important properties of things in the real world. However, they are quite difficult to describe in well-defined terms. The problem of the ‘Definition of Life’ has therefore perplexed scientists and philosophers for centuries. Modern thought is that trying to generate a formal definition of life is futile (Chyba & McDonald Reference Chyba and McDonald1995; Bedau Reference Bedau2010; Benner Reference Benner2010; Machery Reference Machery2012). However, we want to find life (whatever it is) elsewhere, and understand how life (whatever it is) originated. So, what are our experiments to detect if we want to detect primitive or alien life but we cannot define what it is that we are detecting?
This paper does not provide a complete overview of the ‘definition of life’ literature, which has been ably conducted elsewhere (see for example (Chyba & McDonald Reference Chyba and McDonald1995; Anon (editorial) 2007; Kolb Reference Kolb2007; Tsokolov Reference Tsokolov2009; Benner Reference Benner2010; Tirard et al. Reference Tirard, Morange and Lazcano2010; Leitner & Firneis Reference Leitner and Firneis2011; Machery Reference Machery2012; Trifonov Reference Trifonov2012), and the references for Fig. 1). It is fairly well established that a formal definition of life is not possible, at least in part because all terrestrial life is descended from a common ancestor, and so we effectively only have one example of life from which to generalize (Chyba & McDonald Reference Chyba and McDonald1995; Tsokolov Reference Tsokolov2009; Benner Reference Benner2010; Machery Reference Machery2012). Rather, this paper provides a pragmatic approach to decide how confident we are that something is alive, which is what we need for looking for life elsewhere (Conrad & Nealson Reference Conrad and Nealson2001). This is our ‘constructive belief’ of what life is, the belief that we actually act on (Benner Reference Benner2010). It is therefore also about confidence that our beliefs are correct.

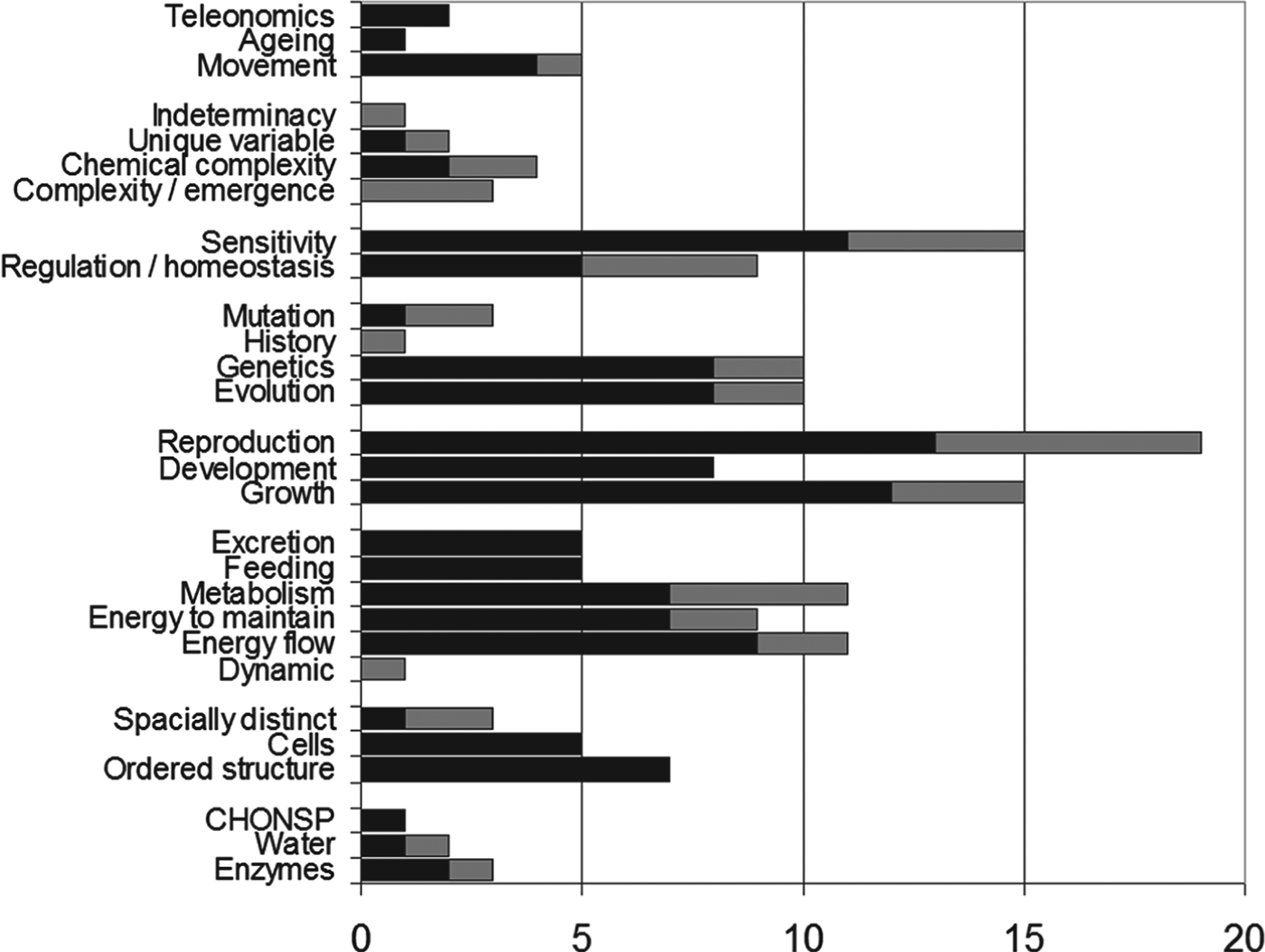
Fig. 1. Textbook definitions of ‘living’. Summary of the characteristics of ‘life’ or ‘living things’ taken from undergraduate textbooks, postgraduate monographs, semi-popular astrobiology texts and some dictionaries of biological terms. As the specific textbook definitions never used exactly the same words, I have translated them into a common vocabulary for comparison. The terms used mean:
• Teleonomics: goal-directed activities
• Ageing: ageing or degeneration with time
• Movement: showing autonomous movement (i.e. not solely moved by external forces)
• Indeterminacy: not linearly predictable, chaotic
• Unique variable individuals:
• Chemical complexity: having very complex (and by implication specific) chemicals
• Complexity/emergence: (usually ill-defined)
• Sensitivity: response to the environment
• Regulation/homoeostasis: active maintenance of an internal state different from the external state
• Mutation: mutation/imperfect transmission of genetic information
• History: biological entities are classified based on their history, not (only) their current properties
• Genetics: possession of a heritable programme or code
• Evolution: evolution adaptation through natural selection
• Reproduction: self-reproduction (i.e. not assembly)
• Development: a programme or progression of change with growth
• Growth: growth by internal incorporation
• Excretion: excreting unwanted material (sometimes includes secretion)
• Feeding: acquiring material from the outside
• Metabolism: active internal conversion chemistry, including respiration
• Energy to maintain: requires energy to maintain structure/integrity
• Energy flow: has energy flowing through it/processed by it
• Dynamic: an active/dynamic/changing/actively maintained system
• Spatially distinct: (other than having cells)
• Cells: organized into cells
• Ordered structure: having an ordered structure (distinct from the environment)
• CHONSP: specification of the chemistry of life as based on C, N, O, P, S and H
• Water: obligatorily based on water
• Enzymes: having highly specific, polymeric catalysts (sometimes specified as proteins)
From general books and textbooks (black bars): (Roberts Reference Roberts1971; Arms & Campbell Reference Arms and Campbell1991; Gould & Keeton Reference Gould and Keeton1991; Raven & Johnson Reference Raven and Johnson1992; Thain & Hickman Reference Thain and Hickman1994; Wallace et al. Reference Wallace, Sanders and Ferl1997; Mayr Reference Mayr1998; Kent Reference Kent2000; Roberts et al. Reference Roberts, Reise and Munger2000; Purves et al. Reference Purves, Sadava, Orians and Heller2001; Roberts & Ingram Reference Roberts and Ingram2001; Brooker et al. Reference Brooker, Widmaier, Graham and Stirling2007; Campbell et al. Reference Campbell, Reece, Urry, Cain, Wasserman, Minorsky and Jackson2008; Lawrence Reference Lawrence2008; Gale Reference Gale2009; Irwin & Schulze-Makuch Reference Irwin and Schulze-Makuch2011; Cranford Reference Cranford2012) and (from the early Victorian period) (Hamilton Reference Hamilton1845). From monographs and research literature (grey bars): (Mayr Reference Mayr1982; Eigen Reference Eigen, Murphy and O'Neill1995; Oro Reference Oro and Schopf2002; Luisi Reference Luisi2006; Scharf Reference Scharf2008; Schulze-Makuch & Irvin Reference Schulze-Makuch and Irvin2008; Fry Reference Fry and Bertka2009; Hazen Reference Hazen and Bertka2009; Leitner & Firneis Reference Leitner and Firneis2011).
This paper seeks to extract a simple approach to testing our confidence that something is alive. Starting with a simple observation of how we intuitively identify animals as ‘alive’, I identify four types of properties that we use to identify living things:
1. Structure, that is highly improbable in its environment.
2. Dynamic maintenance of that structure, through activity that is characteristic of the organism.
3. Occurrence of groups of similar organisms that can be distinguished as a natural group.
4. Substrate-independence: living things are determined by an internal code, not (solely) by their external environment.
I find that evolution and reproduction are not practical tests for life. Reproduction is, of course, central to our understanding of life, but we might have to wait decades to observe it, even in micro-organisms. Evolution is also usually unobservable, and is a logically necessary consequence of reproduction of an organism specified by a code.
Humans intuitively understand the concept of ‘living’
Humans have an intuitive understanding of the difference between ‘living’ and ‘non-living’, or between ‘animate’ and ‘inanimate’. It is probably of selective advantage for omnivores to be able to discriminate between potential food or predators and other objects. This intuitive understanding is probably a pre-formed aspect of our brains in dealing with the visible world, one that we have extended into unseen realms with the invention of the microscope. The psychological literature offers a range of examples of patients with brain damage that specifically impairs their ability to describe living things, without impairing their ability to describe other classes of objects (e.g. man-made things, or things that can move) (Warrington & Shallice (Reference Warrington and Shallice1984), reviewed in Tyler & Moss (Reference Tyler and Moss2001)). Behavioural data from other animals also suggests that at least great apes (Boesch Reference Boesch2012) and some other large mammals can clearly distinguish living from non-living, and the living from the now-dead-but-recently-living.
The cognitive deficits in identifying living things found in rare brain-injured patients are usually strongest in visual recognition (Moss et al. Reference Moss, Tyler and Jennings1997; Capitant & Laiacona Reference Capitant and Laiacona2011). Verbal description is less often affected. This may reflect that our verbal ‘definition’ of life is less well grounded in our own, intuitive understanding, a disconnect that certainly fits with our inability to come up with a formal verbal definition of life. For life, it is literally true that we know it when we see it. However, ‘I know it when I see it’ does not help writers, scientists or logicians.
Textbook descriptions of life
Textbooks usually side-step the problem of defining life by listing properties of life, based more or less explicitly on what ‘everyone knows’. There is substantial uniformity among the textbook lists of properties, which is probably only partly due to their being copied from each other. Fig. 1 summarizes some typical definitions from textbooks and monographs. This analysis is different from the lexicon of life produced by Trifonov (Reference Trifonov2012) in that I have tried to interpret concepts into a common vocabulary, rather than compiling words as originally used.
The number of items chosen from the list in Fig. 1 varies from 4 to 10, but usually at least one is selected from genetics, structure, reproduction and metabolic categories. Some lists are very specific to terrestrial life (e.g. Thain & Hickman Reference Thain and Hickman1994; Purves et al. Reference Purves, Sadava, Orians and Heller2001; Deamer Reference Deamer2010) and do not pretend to be able to generalize away from life that is based on DNA, RNA, proteins, ribosomes and similarly general components of terrestrial life. Quite often the selected characteristic is stated as the ‘capability’ to do something. I will return to what we can do about this ‘capability’ concept below.
This pragmatic, ‘I know it when I see it’ approach to defining life has precedent in the Artificial Intelligence field's attempt to describe intelligence. The Turing Test (which Turing called ‘The Imitation Game’) is a commonly cited method for determining if a system has ‘intelligence’. The system is asked questions, and if the interlocutor thinks the system is behaving in a way that they recognize as ‘intelligent’ then it is, effectively, intelligent. What intelligence ‘is’ is sidestepped (Moor Reference Moor2003). A similar approach is used to diagnose mental disease. The standard diagnostic manual for mental illness (the DSM series) does not attempt to have single, binary rules for diseases such as depression or schizophrenia, but rather has a list of characteristics of people suffering these conditions (American Psychiatric Association 2000). If a patient meets most of the conditions, then they are considered to be subjects for treatment for the disease. Whether they are ‘actually’ depressed or schizophrenic is not relevant, because we cannot know.
However, Fig. 1 has a rather large list of criteria. There are common themes, reflected by the groupings in Fig. 1. For example, aspects of metabolism are linked – if you metabolize, then you must ingest and excrete. If you are a dynamic system (i.e. a system actively maintained) then you must have energy input. This still leaves us with a range of features. Which do we actually use when deciding if something is alive?
A simple experiment
To bring out a key of what is understood by ‘a living thing’ I have carried out the following simple experiment at three scientific conferences, including the recent ASB Conference in Edinburgh, UK in April 2013, and at several smaller seminarsFootnote 1. I asked the audience of the conference to select an observational subject which is not someone well known to them. (This phrasing biases conference audiences to pick someone in the audience, rather than furniture, although ASB5 was remarkably un-biased.) I then tell them I will ask three questions, and want them to indicate, by show of hands, whether the answer is ‘Definitely yes’, ‘Definitely no’ or ‘Don't know/not sure’. The three questions are:
1. Is your subject alive?
2. For those who answered ‘yes’ to question 1: Is your subject human?
3. For those who answered ‘yes’ to question 2: Is your subject vegetarian?
My expectation was well borne out in all the tests – audiences could confidently say that people who they had never seen before were alive, and were human, but could not say whether they were vegetarian.
This is meant as an illustration and the springboard for discussion, and is not in itself a thorough exploration of our understanding of life. The way the question is phrased means that the audience does not explore anything smaller than they can see, or larger than will fit into a large room. The systems where our intuition about the nature of life falters are often very small (viruses) or very large (Gaia), or operate on very different timescales from a conference lecture. However, this is a deliberate aspect of the question. I wished to start from a clear example of living versus non-living, and develop the answers from that example into more general observations.
Below, I will use this simple example as the starting point for a discussion of what the audience was using as criteria for their identification of living humans. I will infer that the audiences were using four characteristics to answer the questions. They are:
1. highly distinctive structure (physical or chemical)
2. dynamic behaviour (physical or chemical)
3. multiple instances of life forming a ‘natural group’
4. independence of structure and behaviour of the details of the substrate on which the life is growing.
The concept of Natural Groups is rarely included in the usual descriptions of the properties of life. Other properties are directly listed or may be indirectly inferred from the characteristics in Fig. 1.
Criteria used to discriminate living from non-living
Structure
Firstly, the most obvious characteristic of life is a complex, distinctive macroscopic structure. It has been amply demonstrated that physical structure alone cannot determine what is alive and what is not (for an astrobiological example, see the discussion on the meteorite ALH84001 (McKay et al. Reference McKay, Gibson, Thomas-Keprta, Vali, Romanek, Clemett, Chillier, Maechling and Zare1996; Thomas-Keprta et al. Reference Thomas-Keprta, Bazylinski, Kirschvink, Clemett, McKay, Wentworth, Vali, Gibson and Romanek2000; Golden et al. Reference Golden, Ming, Morris, Brearley, Lauer, Treiman, Zolensky, Schwandt, Lofgren and McKay2004)). The way we discriminate humans from non-living things is the statistical unlikelihood that a human anatomy forms by what we understand of non-biological processes. However, the smaller and simpler the structure, the less convincing this argument is. A small round blob can be generated by many processes. Head, shoulders, knees and toes are harder to create by water flow, precipitation or tectonics, although quite complex ‘fossil’ forms can be mimicked by geology, a point I come back to below. The statistics of physical appearance are therefore hard to validate. Attempts to define what the morphological features of a living thing are usually come back to saying ‘it is like that living thing over there’, which is not helpful from our point of view (although pragmatically, that is what the conference audiences were doing).
Chemical structure is therefore more commonly discussed in the literature on exo-life detection (Conrad & Nealson Reference Conrad and Nealson2001; Cady et al. Reference Cady, Farmer, Grotzinnger, Schopf and Steele2003; Cleland & Copley Reference Cleland and Copley2005; Parnell et al. Reference Parnell2007; Davies et al. Reference Davies, Benner, Cleland, Lineweaver, McKay and Wolfe-Simon2009; Benner Reference Benner2010). Chemical fingerprints of life were one of the earliest suggested biosignatures in the modern era of exploring space for life. Lovelock's classic description of how life could be detected (Lovelock Reference Lovelock1965) listed both the search for order (non-random chemical structures or molecular masses) as well as non-equilibrium thermodynamics as equally important signatures for life, and illustrated the experimental discrimination between biological and non-biological samples by showing how random hydrocarbons had a continuous molecular weight distribution whereas biogenic hydrocarbons (wool waxes) had discrete molecular weights. Similarly, Joshua Lederberg's analysis of how to detect life (Lederberg Reference Lederberg1965) focused on the low entropy of finding a few, unexpected chemicals as characteristic of life rather than on thermodynamic disequilibrium. Lederberg's leading example was that life used only one enantiomer of many chemicals, whereas random chemistry would generate racemic mixes. Only later did this work focus on thermodynamic disequilibrium as characteristic of life (e.g. Hitchcock & Lovelock Reference Hitchcock and Lovelock1967; Lovelock Reference Lovelock1975).
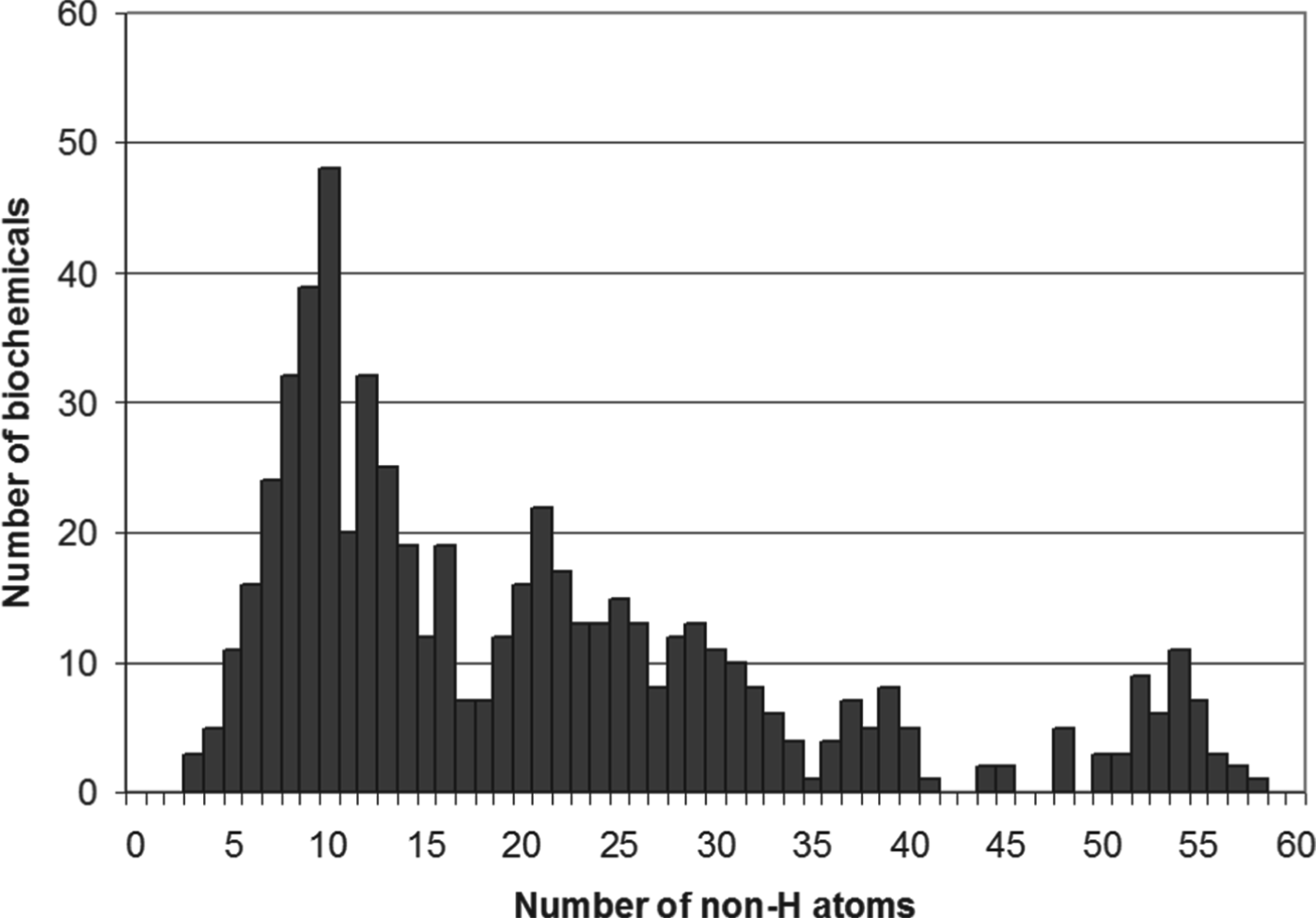
The chances that a specific class of molecules may be present out of a wider chemical space of equally likely, thermodynamically equivalent molecules (e.g. alpha amino acids but not beta amino acids) can be calculated from knowledge of the chemical space of all possible compounds (Bains & Seager Reference Bains and Seager2012), and hence a number put on the concept of ‘improbable’. As a numerical example of how this could be approached, we can consider the molecules of primary intermediary metabolism (ExPasy 2010), which contains around 600 molecules. The size distribution of those molecules (Fig. 2) shows that the majority contain around 10–11 atoms (not counting hydrogens), or are made of modules containing 10 or 11 atoms (the peaks in Fig. 2 at 20, 30 and 40 atoms). If we take 10 atoms as a typical size of a ‘basic metabolite’, there are ∼3.5×107 possible 10-atom molecules made of C, N, O, S(II) and P(V) (Bains & Seager Reference Bains and Seager2012), from which life has selected ∼600, a selection of 1 in 1.7×105. This is an estimate of the probability that we see the same group of 600 chemicals in a wide range of living organisms, rather than either a random mixture of all the ∼3.5×107 possible chemicals, or different random subsets of 600 chemicals in each organism. Although this is a very crude approximation (it ignores the thermodynamic stability of the actual and potential metabolites, for example), it may have some relevance: the information content necessary to select one molecule from a pool of 1.7×105 equates to an entropy of –91 J mole−1, and hence a free-energy requirement of ∼27 kJ mol−1 at 20 °C (see Hitchcock & Lovelock (Reference Hitchcock and Lovelock1967) for a general discussion of the relationship of probability, Boltzman entropy and free energy). Perhaps by coincidence, this is similar to the free energy of hydrolysis of the near-universal energy intermediary in that central metabolism, Mg.ATP (24 kJ mol−1 under physiological conditions (Metzler & Metzler Reference Metzler and Metzler2001)).

Fig. 2. Size distribution of metabolites. Y-axis: number of biochemicals in Expasy metabolic map of ‘core’ metabolism. X-axis: number of non-hydrogen atoms in biochemicals. Data source (ExPasy 2010; Bains & Seager Reference Bains and Seager2012).
For our purposes, then, a highly unexpected macroscopic structure (eyes+ears+mouth+nose) or a highly statistically improbable chemical structure (sterols, DNA) is a signature of life.
Of course, having a group of similarly structured objects does not prove they are living. A defined, unexpected, statistically improbable structure gives us confidence that we are looking at an example of life, but it is neither necessary nor sufficient to prove the presence of life.
Dynamic
The second feature is that life is dynamic. Life's dynamic nature is not explicitly explored in the questions above. (This could be explored by adding a fourth question ‘is your subject dead?’, but this overlaps with the first question.) However, empirically we know when someone is alive versus dead because they move, breath and ask questions. Indeed, in the Christian bible, ‘life’ and ‘breath’ are almost synonymous.
The requirement that life is dynamic fits with the consensus that life needs energy (Lineweaver & Egan Reference Lineweaver and Egan2008), and indeed the ‘follow the energy’ strategy for identifying habitats has substantial success (Hoehler et al. Reference Hoehler, Amend and Shock2007). However, I chose to state that life is dynamic rather than life needs energy, because our intuitive recognition of life is based on its dynamic character, not on the mechanism by which that dynamic character is maintained. Again, I emphasize that my purpose is to identify those aspects of our intuitive understanding of life that can be used as practical tests for life. What we observe is dynamic behaviour, what we infer is energy dissipation.
Schrödinger's comment that life ‘feeds on negative entropy’ (Schrödinger Reference Schrödinger1944) is related to this energy requirement, as gain in entropy is thermodynamically equivalent to loss of free energy: indeed the statement about entropy is more helpfully stated by saying that life ‘feeds on free energy’, and Schrödinger would probably have put it this way if he had been a chemist. This is different from the discussion above about life being characterized by improbable structures (and hence low internal entropy): life can generate low internal entropy by capturing and exploiting a wide range of reactions (reviewed in Seager et al. (Reference Seager, Schrenk and Bains2012)) which are mostly driven by enthalpy changes (specifically redox changes), not entropy changes. The observation that life needed chemical energy was well established by the time Schrödinger arrived at it – see for example Baas-Becking & Parks (Reference Baas-Becking and Parks1927), McLean (Reference McLean1938) and Winzler & Baumberger (Reference Winzler and Baumberger1938). I also note that ‘What is Life?’ did not attempt to answer its own question, and the question it did answer – ‘What is a gene?’ – had been addressed experimentally by biologists by the time Schrödinger was speculating about it (Avery et al. Reference Avery, MacLeod and McCarty1944), although it was another decade before we had an understanding of why nucleic acid was the chemical that carried genetic information.
Again, dynamics is not sufficient to describe life. This is the error that Lineweaver & Egan (Reference Lineweaver and Egan2008) make – hurricanes are dynamic systems that dissipate external energy gradients, but are not what we understand as ‘alive’, for reasons captured in the next two sections – ‘Natural groups’ and ‘Independence of substrate’.
Natural groups
Thirdly, life forms Natural Groups. The conference audiences could immediately recognize humans among the millions of species present on the Earth. This is because we observe that living things form ‘Natural Groups’ or ‘Natural Kinds’. There is a statistically robust difference between the world's population of elephants and the world's population of bison, at anatomical, behavioural and genetic levels. In terms of numbers of horns, tusks and trunks, size of ears and teeth, elephants and bison form two non-overlapping sets of entities, even though in colour, mass, number of legs and eyes they may overlap. Elephants form a ‘Natural Group’ (Bedau Reference Bedau2010), i.e. a group of objects that can be objectively distinguished from another group of objects with no intermediate stages between them, as do bison, mice, and (as illustrated by the answers from conference audiences to the questions put to them) humans. If I asked audiences to distinguish a human from a gorilla or a chimpanzee, even a Homo sapiens sapiens from Homo sapiens neaderthalis they could have readily done so. In contrast, if we asked them to tell whether Arthur's SeatFootnote 2 was a hill or a mountain, they would consider that its height was probably the relevant primary distinguishing feature between hills and mountains, and make a decision based on its height. However, there is no clear partition of geographical features into hills and mountains based on height, or any other characteristic. The threshold between them is arbitrary. ‘Hills’ and ‘Mountains’ are not Natural Groups in the same way that ‘Humans’ and ‘Chimpanzees’ are natural groups, distinct in a range of characteristics. Of course, there is no clear distinction between a human and a chimpanzee if you judge them solely on height. However, there are wealth of other anatomical and genetic differences that can uniquely discriminate between the two species.
I emphasize that finding a Natural Group does not guarantee that the class of objects concerned is alive. Cars are not alive despite falling into well-defined, easily identifiable structural categories (Steele & Toporski Reference Steele and Toporski2010). Occurrence of Natural Groups is one of four criteria, none of which are necessary or sufficient to identify living things (although the presence of cars is a strong biosignature on Earth: they would not be here if the Earth was not inhabited).
A necessary component of the Natural Group criterion is that we can define the physical boundaries of the living thing, otherwise we cannot count them. The need for a boundary around a living thing is often stated in terms of a semi-permeable boundary or membrane (Luisi Reference Luisi1998). Boundaries around cells are necessary to keep the cell's contents together, and provide barriers across which gradients can develop. Organismal boundaries may be necessary for an organism to maintain an organized structure and a high internal free energy state etc.. For practical purposes, however, we use the fact that living things must have a distinct physical boundary to identify them as distinct entities, and hence distinct members of a Natural Group.
The existence of Natural Groups is the observed outcome of reproduction and genetics. Inheritance is central to our understanding of what life is (Benner Reference Benner2010; Tirard et al. Reference Tirard, Morange and Lazcano2010): properly understood history is evidence for genetics, not visa versa. Humans inherit humanity from their parents. However, we can rarely see this happening – certainly in a conference auditorium the amount of reproduction going on is small. However, its consequences are entities with tightly limited collections of properties. In contrast, a hill inherits its characteristics (height, geology, slope steepness, etc) from its parent mountain continuously. A small change in any aspect of the hill causes a small change in the properties of the whole. There is no equivalent of a single base change turning a wing into a leg in Drosophila. A glacier will erode a mountain into a hill one boulder at a time.
A classification into Natural Groups breaks down in some cases such as ring species (reviewed in Irwin et al. (Reference Irwin, Irwin, Price, Hendry and Kinnison2002)), and in evolutionary time: indeed ring species are rare spatial examples of a continuum of change that occurs more commonly in species separated in time (Dawkins Reference Dawkins2004). When asking for life to fall into Natural Groups, we ask about what we can see here, now, not what we could find if we explored far enough in time and space.
Implicit in a description of Natural Groups is that there is more than one example of the life to be found. Most new species on the Earth are found initially as a single specimen: they are singletons (and hence usually the type specimen) of their species. Are we to conclude that they are not alive until we find another example? I will discuss why we would not take this extreme position below.
Independence of substrate
Fourth and last, we know from experience and can observe directly that living things can maintain their structure and their characteristic features that are statistically distinct from other living things on a variety of substrates (foodstuffs). This was the focus of my questions to the conference audience, and brought out a central difference between structured, dynamic things that are living and things that are not alive. My conference audience could not tell whether a stranger ate meat or cereal for breakfast. A pig eats sausage and turn it into a pig, whereas a human eats sausage and turns it into human. In contrast, the flame that takes a candle and turns it into CO2 and H2O looks like a candle flame, whereas the flame that burns a match looks like a match flame, even though the candle was lit by a match. For the candle, the substrate determines the physics and the chemistry of these dynamic, evolving systems. A human who eats a candle continues to look human.
This has an inverse implication – that a living organism growing on different substrates will generate different waste products, i.e. will chose to ‘throw away’ different components of ingested material in order to maintain a (nearly) constant internal structure and chemistry. Thus, a bacterium fed solely amino acids will generate nitrogen-containing waste (probably ammonia), whereas one fed glucose will not excrete ammonia. The output of life is as varied as the input, but the internal structure remains largely the same. A bacterium growing on nitrogen-rich medium is not identical to one grown on nitrogen-poor medium. Different sets of enzymes are induced and repressed in the two cases, and different growth rates may result in different fractions of cell mass represented by ribosomes, DNA, cell wall and other components. However, the organism will be clearly identified as the same Natural Kind despite differing substrates, because those features that distinguish it from other organisms will remain essentially the same. In the case of bacteria this may be DNA sequence, in the case of humans anatomy and genetics.
This is related to deeper concepts of ‘autopoiesis’ – making yourself (Margulis & Sagan Reference Margulis and Sagan2000). The key distinguishing feature that conference audiences identified easily is that living things make themselves from a variety of parts.
Both the requirement for Natural Groups and the requirement for Independence of Substrate require that we see more than one example of a living thing. Thus, asking whether a specific object is ‘alive’ is not that easy. In reality, we put a specific object into context, and ask whether an environment contains living things. When asking the audience to pick an experimental subject in the example above, we are in fact asking them to survey an environment, not analyse an entity isolated in a vacuum.
Critical features derive from coded descriptions
Where is genetics in this? Implicit in all the four features listed above, and many of those in Fig. 1, is that their formation or action is directed by a centrally acting code, which is a key feature of life (Eigen Reference Eigen, Murphy and O'Neill1995; Bains Reference Bains2004).
In particular, substrate independence implies an internal code. Something inside the human dictates what a human should look like, without itself looking like a human, without being influenced by the environment of the human, and that something is not a gross physical or chemical property of the human. What determines how a crystal grows or a flame burns are the gross, overall properties of the system – the crystal facet, the wick and wax. Candles burning and crystals forming are examples of pattern-directed propagation. In contrast, humans are examples of coded replication.
A code has a one-to-one relationship with what is coded, but it is indirect, not inferable from the code without the translation apparatus. The word ‘haggis’ means a haggis, but it does not look or taste or sound like a haggis. There is no reason for ‘haggis’ not to mean a member of the Bolivian taxi drivers union, rather than a thick sausage made from oats and offal stuffed in a sheep's stomachFootnote 3. A cook cannot take the word ‘haggis’, drop it into a kitchen and expect a haggis to spontaneously crystallize from the meat locker. Less facetiously, she cannot say to another chef ‘make me a haggis’ and have any chance of getting what she was expecting without also providing a translation, i.e. a recipe. If I misspelled ‘haggis’ as ‘higgs’ and asked someone to make me a higgs, the result would be a 27 km particle accelerator rather than a 500 g sausage. A small change in the code results in a large change in what is coded, both in structure and function, because the map from code to structure is arbitrary. In contrast, pattern-based propagation is driven directly from the pattern: a crystal grows to copy the crystal facets of the seed. Dislocations grow to copy the dislocation. Pattern-based propagation is limited by physics or chemistry to a very few options: pulverizing a crystal to nano-particles and using these to seed crystal growth will result in the same crystal, every time (polymorphs excepted). Code-based propagation is only limited by what is physically possible.
Benner has made an equivalent point in a chemical context, when stating that polyelectrolytes are optimal genetic material (Benner et al. Reference Benner, Ricardo and Carrigan2004) because you can change them arbitrarily and still have almost identical chemical function: it is this sameness of chemistry that allows them to work as the genetic material. The chemistry of replication of DNA is almost completely unaffected by what it codes for (Benner et al. Reference Benner, Ricardo and Carrigan2004). DNA does not provide a pattern or a template for an organism, but a coded description, and so its replication must be as unaffected by its coded content as possible. In contrast, the various ‘self-replicating’ RNA, protein and other chemical systems proposed as precursors for life are patterned replication. If you change the chemical pattern it no longer replicates itself.
Evolution is a direct consequence of self-reproduction of a code (Eigen Reference Eigen, Murphy and O'Neill1995). Transmitting an arbitrary message takes energy; higher the accuracy required, more the energy. Unless infinite energy is expended, changes, i.e. mutations, will creep in when the code is copied. There is an information theoretic argument for this, but familiarity leads me to prefer the thermodynamic one. When adding any monomer to a polymer via a coded mechanism, there is a choice between inserting the correct monomer and the incorrect one. In DNA synthesis, a range of energy-dissipating mechanisms drive the choice towards the correct base, with extraordinary fidelity in animal DNA polymerases (Goodman Reference Goodman2002). However, it is still in effect a chemical equilibrium. The relationship between the free energy of a reaction (ΔG) and the equilibrium constant of a reaction (K)
(where R is the gas constant and T is the absolute temperature) says that the only way that an equilibrium can be 100% on one side of an equation, i.e 100% fidelity for the correct monomer is when ΔG=−∞. Hence, perfect replication of a code is not possible, mutations will occur, and as a necessary result a population of organisms that share a common ancestor will contain members with different genes, and hence potentially different phenotypes. Malthusian logic dictates that the number of potential organisms is greater than the number that can actually be accommodated by any finite environment. As a result, there will be competition, and those whose phenotypes are better fitted to win that competition will therefore surviveFootnote 4. The comment by Deamer (Reference Deamer2010) ‘Suppose the system reproduced perfectly so that evolution could not occur. Would it still be considered to be alive?’ is therefore physically implausible. Equally, ‘mortality’ is not a basic descriptor of life (Bedau Reference Bedau2010) – it is an inevitable consequence of the eventual failure of propagation of coded information as an organism grows and self-repairs (Kirkwood & Holliday Reference Kirkwood and Holliday1979). It is only a matter of how much energy the organism is willing to spend to stay alive that holds this failure at bay, but infinite energy is not a realistic option.
This arbitrary nature of a code also links to another concept that has followers in the ‘what is life’ argument, that of complexity and emergence. Emergence is as hard to define as life, but Ronald et al. (Reference Ronald, Sipper and Capcarrere1999) conclude that a central feature is a surprise – the observer or experimenter is surprised that a system produced the result that it did. A result emerged, unexpectedly, from simpler, apparently predictable properties. This is the essence of genetics, where a single base pair change in 1.2×108 bases of Drosophila genome can change a wing into a leg, but many other single base pair changes apparently do nothing at all. The highly non-linear mapping from code to organism results in surprise all the time.
Finally, a coded description explains why we can distinguish organisms based on Natural Groups. If the organism is programmed by its coded instructions, and any coded instructions are equally valid (albeit maybe teaching the formation of a non-viable organism), then the number of possible organisms is vast. If the genome size of an organism is (say) 5×106 bases, there are ![]() $4^{5 \times 10^6} $ possible genomes for that organism, a staggeringly large number that, if implemented in the tiny SAR11 cells (Rappe et al. Reference Rappe, Connon, Vergin and Giovannoni2002) at one genome per cell, would require a ball of cells 101000000 light years in radius. Obviously, we cannot experience that variation. The actual organisms we do experience are a tiny, arbitrary sampling of the phase space of possible organisms, with the chance of any two organisms being indistinguishable in all features being essentially zero unless they are related by descent. Thus, organisms are distinct. They inherit their code (including some mutations), and so a group of organisms sharing a common descent look the same, and different from other organisms. In more commonplace terms, humans look like humans because of their human genes, and all humans look like humans because they inherited human genes.
$4^{5 \times 10^6} $ possible genomes for that organism, a staggeringly large number that, if implemented in the tiny SAR11 cells (Rappe et al. Reference Rappe, Connon, Vergin and Giovannoni2002) at one genome per cell, would require a ball of cells 101000000 light years in radius. Obviously, we cannot experience that variation. The actual organisms we do experience are a tiny, arbitrary sampling of the phase space of possible organisms, with the chance of any two organisms being indistinguishable in all features being essentially zero unless they are related by descent. Thus, organisms are distinct. They inherit their code (including some mutations), and so a group of organisms sharing a common descent look the same, and different from other organisms. In more commonplace terms, humans look like humans because of their human genes, and all humans look like humans because they inherited human genes.
Reproduction, evolution and the NASA definition of life
Does this mean that reproduction, genetics and evolution are key descriptors for life, as suggested by the NASA definition? There has been a tension between definitions of life based on metabolism and those based on evolution (Nealson & Conrad Reference Nealson and Conrad1999), with evolutionary definitions taking the lead following NASA's definition of life, which is commonly cited as
‘Life is a self-sustained chemical system capable of undergoing Darwinian evolution’
(Luisi Reference Luisi1998). Several textbooks also suggest that evolution is a useful descriptor for life, but unfortunately NASA and the books are wrong.
Including evolution in a central description of life is not of any practical use. It does not lead to measurable parameters (Leitner & Firneis Reference Leitner and Firneis2011) because, as Luisi (Reference Luisi2006) has pointed out, evolution happens in populations, not individuals, so you have to study enormous amounts of life to see it happening, and it happens slowly, so you have to watch that life for very long periods of time. Fast-growing, short-lived species can be observed to evolve in the laboratory (reviewed in Burke et al. Reference Burke, Dunham, Shahrestani, Thirnton, Rose and Long2010; Conrad et al. Reference Conrad, Leris and Palsson2011; Kawecki et al. Reference Kawecki, Lenski, Ebert, Hollis, Olivieri and Whitlock2012), and in the field. However, during the time needed to observe evolution you will see the life doing everything else listed in Fig. 1: grow, metabolize, reproduce, etc. The ‘evolution’ tag adds nothing.
The ‘evolution’ tag also does not add to our understanding of life. As argued above, any coded self-replicating system must undergo evolution as a logical necessity. Saying that life is something that undergoes Darwinian evolution just replaces a potentially observable description of coded replication with an unobservable consequence of coded replication.
In reality, we often do not watch evolution happening, we infer it has happened from modern homologies and from the fossil record. Thus, we infer that a species has the ‘capability’ to evolve from its past evolution (and from structure – I will return to this below). The evolution of humans is of consummate interest, but we cannot see it happening, so we infer that evolution has happened from comparison of present and past life, identified from the fossil record (and from DNA, but DNA is an archetypical example of the chemical structure that we use to identify life, as discussed in the section on Structure above). However, fossils are usually identified by their homology with living forms. The more disparate from living forms a putative fossil is, the more the paleontologist falls back on the criteria above, of complex (non-random) structure, Natural Groups, and (to the extent that this can be extrapolated) independence of substrate. Thus, fossil dinosaurs were immediately recognized as an extinct form of life, whereas there was debate for some years whether the simpler Ediacaran body forms were really living at all, only resolved when more complete fossils showed complex body plans (the argument from unlikely structural forms) and more fossils were found (the argument from Natural Groups) (See Narbonne (Reference Narbonne2005) for a review of the Ediacaran biota). The claims that even earlier trace fossils show that animal life was present 1000 My before the present (Seilacher et al. Reference Seilacher, Bose and Pflüger1998; Rasmussen et al. Reference Rasmussen, Bengtson, Fletcher and McNaughton2002) are still controversial, because there are few traces, and they are not clearly distinct from structures that could be formed from unicellular life (Seilacher et al. Reference Seilacher, Bose and Pflüger1998; Morris Reference Morris2002). Thus, in practice, we show that fossils are traces of ancient life either by clear homology with modern life (an argument from Natural Groups) or by applying all the criteria listed above, in the Section – Criteria used to discriminate living from non-living.
Even reproduction is not that useful in identifying life, no matter that it is a key feature of life. This strikes at the heart of the ‘capability’ argument. We know that many micro-organisms cannot be cultivated, i.e. cannot be observed to reproduce, in the laboratory (Nealson Reference Nealson2009). Does that mean that 95% of bacteria are not alive? This seems implausible. We assume that some at least have the capability to reproduce, but we infer this because we see Natural Groups of highly chemically improbable structures (usually DNA sequences) in the soil. Reproduction, or the past capacity to reproduce, is inferred from the presence of distinct structure, Natural Groups, and present active metabolism.
Even for macrofauna, it has taken decades of research to find out how to breed some mammals in captivity – were the Sumatran rhinoceros or the Giant Panda considered not alive before being observed to breed in captivity? Obviously not: again, their breeding was inferred from the presence of more than one rhinoceros or panda in a diverse set of environments, and the lack of correlation of panda or rhinoceros with local geochemical sources of their components, and the fact that they were distinct Natural Groups (i.e. species). Many individual organisms cannot reproduce. Humans cannot reproduce at all – a pair of humans can produce a new human, but one human cannot reproduce (see Benner (Reference Benner2010) for more on this). Rather, we infer reproduction by the presence of over 7 billion humans on Earth.
As a description of a feature of life, reproduction is central (unlike evolution, which is a derived feature), but as an observational criterion it is not effective, and is better replaced by the inevitable, and observable, results of reproduction.
Tests for life
Summarizing the arguments above, our practical tests for life are:
1. Structure: A living thing has a structure that is highly improbable in its environment, which in chemical terms means out of thermodynamic equilibrium with its environment but also made of a systematic subset of possible chemicals, i.e. with very low entropy as well as high enthalpy.
2. Dynamic: A living thing maintains itself dynamically, ‘feeding on’ its environment. The pattern or nature of its activity (physical or chemical) is characteristic of the organism.
3. Groups: Living things do not come in isolation – there are always many of them that can be distinguished as a natural group. In physical terms this means that a living thing must be spatially bounded (as otherwise you cannot count it/them).
4. Substrate-independent: A living thing maintains this specific pattern of structure and activity in a variety of conditions. If the conditions do not allow the living thing to be alive, then it will not be alive rather than change to another living thing.
Evolution and reproduction are not included in this list as practical tests for life. Reproduction is central to our understanding of life, but we might have to wait decades to observe it, even in micro-organisms. Evolution is an inevitable consequence of reproduction and thermodynamics, and is even harder to observe.
The requirement to find multiple living things, and not just one, might seem overly harsh. What if we find one bandersnatch on Mars: will we say that there is no life on Mars until we find a second bandersnatch? If we found one bandersnatch, and many other, similar creatures, then we would have a Natural Group, albeit one with more variation within it than we are used to on Earth. If the only potentially living thing on the planet was one bandersnatch, then formally, we should be skeptical that it is alive. Until we see several of them, we really cannot be sure. However, if we did find a bandersnatch on Mars, the other three criteria would be met in full, as it would have complex, highly improbable molecular and anatomical structure, it would be dynamic, and it would not be obviously derived from its substrate (Carol Reference Carol1876). We would therefore be confident that there was life on Mars. (The same argument applies to finding a single new member of a potential new species on Earth.) The example emphasizes what the list above is, and what it is not. It is a list of criteria derived from how we recognize life, and as such each of the elements gives us confidence that an observed object is alive. One could derive statistical measures for each of the criteria, in the fashion that I have illustrated in the section on Structure above, and aggregate them to a statistical estimate of how confident we were that a subject was alive. What it is not is a formal definition, where each element has to be met. As stated at the start of this paper, that formal definition is not my goal, and probably is not possible.
This is not a particularly original list. It is similar to the ‘Programme–Metabolism–Container’ description of life (Bedau Reference Bedau2010). However, it focuses on things that are measurable, as summarized in the last section.
Practical detection of life
Based on the list of criteria above, we can describe specific tests for life that address each element of the evidence. Note again that each test adds to our certainty, but on their own are not proof or disproof of the presence of a living thing.
Physical structure can be detected by examination, but as has been shown in the ALH84001 example, simple structures are not a reliable indicator of biogenesisFootnote 5. Thus, complex macroscopic structures (humans, trees and mushrooms) are strong evidence of life, but microscopic structures may remain debatable. For this reason, chemical structure is a preferred route, as discussed above. This is a chemical analytical task. Specifically, it should look for subsets of equivalent molecules, such as alpha versus beta amino acids (Lu & Freeland Reference Lu and Freeland2008), mono-halogenated versus multiply halogenated hydrocarbons (Biemann et al. Reference Biemann, Oro, Orgel, Neir, Anderson, Simmonds, Flory, Diaz, Rushnek and Biller1976, Reference Biemann1977; Bains Reference Bains2013), or compounds with highly biased chirality (Levin Reference Levin2009; Sun et al. Reference Sun, Saccomanno, Hedlund and McKay2009; Warmflash et al. Reference Warmflash, Chu, Siefert and Fox2009).
Dynamics can be observed macroscopically or chemically. Clearly non-random, powered movement on any scale is a suggestion of life. (Carl Sagan captured the macroscopic aspects of both dynamics and structure when he commented, when asked how the Viking mission scientists would know if they had found life on Mars, ‘If a herd of elephants stampeded across the field in front of the camera, we would not have any doubt about the existence of life on the Red Planet.’ cited in Oro (Reference Oro and Schopf2002)). However, not all life shows obvious physical movement. The output of chemical dynamics may be easier to detect and more universal, but can be confused with equivalent geochemical processes (discussed in Seager et al. (Reference Seager, Schrenk and Bains2012)).
Dynamics and substrate independence can be demonstrated chemically by showing that the same chemical features are generated (i.e. grow) when fed different substrates, or that the putatively living object generates different waste products from only subtly different substrates (Davies et al. Reference Davies, Benner, Cleland, Lineweaver, McKay and Wolfe-Simon2009). Again, alpha versus beta amino acids, ethanol versus propanol, methylamine versus ammonia and compounds of different chirality (Levin Reference Levin2009; Sun et al. Reference Sun, Saccomanno, Hedlund and McKay2009; Warmflash et al. Reference Warmflash, Chu, Siefert and Fox2009) are potential discriminators.
Natural Groups then simply require that we see the same effects several times.
This is not an original list. Indeed, it is not much different from life detection strategies suggested in the 1960s (Lederberg Reference Lederberg1965; Lovelock Reference Lovelock1965, Reference Lovelock1975; Hitchcock & Lovelock Reference Hitchcock and Lovelock1967; Sagan Reference Sagan1975). The point of revisiting such well-trampled ground is to demonstrate that these tests are based on what we can do, they are based on our intuitive understanding of what life is and not a (probably futile) attempt at formal definition, and they do not include reference to unobservables such as reproduction or genetics, although they are based soundly on the presence of both, nor on evolution. It is my hope that this rather pragmatic approach will be of some help to designing experiments and missions, even if it does not satisfy our philosophical longings for a ‘definition’ of life.
Acknowledgements
I am grateful to Dirk Schulze-Makuch (Washington State University) and Andrew Clarke (British Antarctic Survey, Cambridge) for insightful comments, to Jane Garrison (Department of Psychology, Cambridge, UK) for references to the psychology of our understanding of what is living and to the two referees who made many helpful and insightful comments that helped to improve this paper substantially. I am very grateful to Carl and Barbara Berke for supporting me during this work, and to Sara Seager (EAPS, MIT, MA) for continued encouragement and support.