Introduction
The possibility of Mars harbouring life has been a fascinating topic for several centuries, and in recent decades spacecrafts have been used to investigate the past or present habitability of the Red Planet. In 1976 the Viking landers did not detect any organic compounds (representative of life on the surface) above the detection limit of a few parts per billion (ppb) (Biemann & Lavoie, Reference Biemann and Lavoie1979). This was explained by the strong UV radiation and oxidizing conditions of present-day Mars, which would destroy organic compounds (Klein, Reference Klein1978; Benner et al. Reference Benner, Devine, Mateeva and Powell2000; Squyres et al. Reference Squyres, Grotzinger, Arvidson, Bell, Calvin, Christensen, Clark, Crisp, Farrand and Herkenhoff2004). However, when buried and shielded from exterior radiation, organic compounds are capable of surviving for geological periods of time (Kminek & Bada, Reference Kminek and Bada2006; Aubrey et al. Reference Aubrey, Cleaves, Chalmers, Skelley, Mathies, Grunthaner, Ehrenfreund and Bada2006; Kanavarioti & Mancinelli, Reference Kanavarioti and Mancinelli1990). As large amounts of carbonaceous material are thought to be delivered to the surface of Mars by interplanetary dust particles (IDPs) and meteorites every year (Chyba & Sagan, Reference Chyba and Sagan1992; Zent & Mckay, Reference Zent and McKay1994; Flynn, Reference Flynn1996; Bland & Smith, Reference Bland and Smith2000), some residual extraterrestrial organic compounds would be expected on Mars. Hence, the search for both biotic and abiotic sources of organic matter continues. In order to help prepare for future life-detection missions (such as the Mars Science Laboratory and ExoMars missions), it is necessary to perform extensive fieldwork in terrestrial locations that resemble Mars (for a review, see Marlow et al. 2010). As the Martian regolith is extremely dry and the surface is depleted of organic compounds (Biemann et al. Reference Biemann, Oro, Toulmin, III, Orgel, Nier, Anderson, Simmonds, Flory, Diaz and Rushneck1976, Reference Biemann, Oro, Toulmin, Orgel, Nier, Anderson, Simmonds, Flory, Diaz and Rushneck1977; Biemann, Reference Biemann2007), terrestrial deserts have been used as Martian analogue locations. Analyses to detect organic compounds were performed in soils from the Atacama and Arequipa deserts (Skelley et al. Reference Skelley, Scherer, Aubrey, Grover, Ivester, Ehrenfreund, Grunthaner, Bada and Mathies2005; Buch et al. Reference Buch, Glavin, Sternberg, Szopa, Rodier, Navarro-González, Raulin, Cabane and Mahaffy2006; Peeters et al. Reference Peeters, Quinn, Martins, Sephton, Becker, van Loosdrecht, Brucato, Grunthaner and Ehrenfreund2009), as these are two of the driest and most inhospitable places on Earth for microbial life (Navarro-González et al. Reference Navarro-González, Rainey, Molina, Bagaley, Hollen, de la Rosa, Small, Quinn, Grunthaner and Cáceres2003; Gómez-Silva et al. Reference Gómez-Silva, Rainey, Warren-Rhodes, McKay, Navarro-González, Dion, Nautiyal and Varma2008). The present study continues the analyses of organic compounds in Martian analogue soils, and was performed on soils collected near the Mars Desert Research Station (MDRS) in the Utah desert, which receives 140 mm of annual average precipitation (Godfrey et al. Reference Godfrey, Everitt and Martin Duque2008). The Utah soil displays mineralogies similar to Mars (Borst et al. Reference Borst, Peters, Foing, Stoker, Wendt, Gross, Zavaleta, Sarrazin, Blake and Ehrenfreund2010), with sedimentary deposits of sands, evaporites, clays and gypsum (Kotler et al. Reference Kotler2011). A good understanding of the interactions between the organic, mineralogical and microbial components of desert soils is crucial to determine the best locations for future space missions to Mars to be able to detect organic compounds indicative of life. Therefore, we have measured the amino acid content of Martian analogue soil samples, and related these results to the microbial and mineralogical data of the soil samples.
Materials and methods
Samples
Soil samples were collected in the period between 17 and 27 February during the 2009 EuroGeoMars campaign (Foing et al. Reference Foing, Stoker, Zavaleta, Ehrenfreund, Direito, Kotler, Martins, Orzechowska and Thiel2011) from different locations near the MDRS in the south-east area of Utah (USA) (Fig. 1, Table 1). Shovels and spatulas used to collect the soil samples were sterilized by rinsing with ethanol (70%, v/v). Soil samples were then stored in sterile low-density polyethylene bags prior to amino acid analysis in the laboratory at Imperial College London. Soil samples came from several geological formations (Table 1) (Chronic Reference Chronic1990; Fillmore Reference Fillmore2000). Soil samples P1, P8, P10 and P11 are from the Mancos Shale Formation (sample P1 belongs to the Tununk Member), which was formed during the Cretaceous Period with records showing falling and rising cycles of sea level. This formation is very rich in sulphides, with some locations containing gypsum. Samples P2 to P7 are from the Morrison Formation, belonging to the late Jurassic Period. This formation contains bentonite-rich shales deposited in mudflats, with sparse units of fine-grained sandstones. Samples P13 and P14 are from the Dakota Sandstone Formation and were formed during the Cretaceous Period. They were collected from locations that contained fossils of shells in the upper layer. The soil pH and organic matter content (Orzechowska et al. Reference Orzechowska, Kidd, Foing, Kanik, Stoker and Ehrenfreund2011), microbial community content (Direito et al. Reference Direito, Ehrenfreund, Marees, Staats, Foing and Röling2011) and mineral composition (Kotler et al. Reference Kotler2011) were determined. A powdered sample of serpentinite (hydrated magnesium silicate), provided by the Natural History Museum Bern, was used as a procedural blank. It was heated to 500°C for 3 h and subjected to the same amino acid experimental procedure as the soil samples.

Fig. 1. Map showing the locations where the MDRS desert soil samples were collected (south-east area of Utah, USA). The right-corner inset shows the states surrounding the state of Utah (USA). Taken from Direito et al. (Reference Direito, Ehrenfreund, Marees, Staats, Foing and Röling2011).
Table 1. The MDRS desert soil samples (see Fig. 1 for location) and the corresponding coordinates, altitude, depth and formation from which they were collected

Chemicals and tools
All chemicals and tools were purchased from Sigma-Aldrich, except for the AG® 50W-X8 cation exchange resin (100–200 mesh, from Bio-Rad), the 1 ml V-vials (from Fisher) and the trifluoroacetic anhydride/isopropanol (TFAA-IPA) derivatization kit (from Alltech). All the tools, glassware and ceramics used in the amino acid analysis were sterilized by wrapping in aluminium foil and placing in a furnace at 500°C for 3 h.
Amino acid extraction, derivatization and gas chromatography–mass spectrometry (GC-MS) analysis
Around 100 mg of each soil sample was placed inside Pyrex test tubes (16×150 mm) with 1 ml high-performance liquid chromatography (HPLC) grade water, flame sealed and put on a heating block at 100°C for 24 h. After the hot-water extraction, the outsides of the tubes were rinsed with HPLC grade water before being opened; 500 μl of the supernatant of each soil sample was transferred to a smaller test tube (3 ml) and vacuum dried. Each small test tube was placed inside a Pyrex test tube containing 1 ml 6 of M HCl. The Pyrex test tubes were flame sealed and placed in an oven at 150°C for 3 h. After the acid hydrolysis, the outsides of the Pyrex tubes were rinsed with water before being opened. The small test tubes were taken out and the extracts were dried prior to the desalting step. The dried extracts were brought up in 3 ml water, desalted on a cation exchange resin and eluted with 5 ml of 2 M ammonium hydroxide. The eluates were vacuum dried , dissolved in 100 μl of water, transferred to 1 ml V-vials and dried with a flow of N2.
100 μl of an acetylchloride:isopropanol mixture (30:70, v/v) was added, and the vials were tightly capped with Teflon-lined screw caps and placed in a heating block for 1 h at 110°C. The samples were checked 5 min into the reaction to make sure that the vials were tightly capped. After the esterification step, the vials were cooled to room temperature, and the samples were slowly dried with a flow of N2; 100 μl of methylene chloride and 50 μl of TFAA were added. The vials were tightly capped and heated at 100°C for 10 min in a heating block. The vials were cooled to room temperature, and the samples were slowly dried with a flow of N2; 65 μl of methylene chloride and 10 μl of pyrene in methylene chloride (200 μg μl−1), which was used as external standard, were added to the samples; 1 μl of the sample was injected into the GC-MS sysyem.
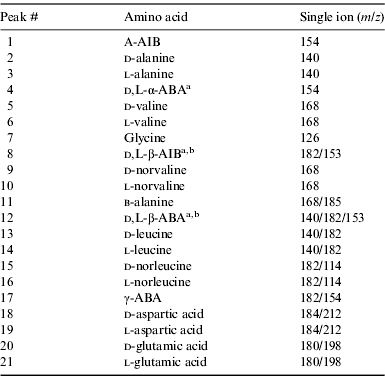
GC-MS analyses were performed using an Agilent 7673 series injector, 6890 network GC system and 5973 mass selective detector. Separation of the d, l-amino acid enantiomers was achieved using a Helifex Chirasil–Val column (50 m×0.25 mm ID×16 μm film thickness) from Alltech. Helium was used as carrier gas with a flow of 1 ml min−1. The injection port (splitless injection) and the MSD transfer line were set to 220°C. The oven programme was held for 5 min at 5°C, increased by 2°C min−1 to 80°C and held for 5 min, increased by 1°C min−1 to 100°C, increased by 2 °C min−1 to 200°C and held for 10 min, and finally increased by 10°C min−1 to 220°C and held for 5 min (total run time of 104.5 min). MS quad was set to 150°C and MS source to 230°C. Data were analysed using the MSD ChemStation Data Analysis software package. Amino acids were identified by comparison of the retention time and mass fragmentation pattern with known amino acid standard mixtures (Table 2 and Fig. 2).

Fig. 2. The 10–70 min region of the single ion GC-MS traces (m/z 126, 140, 154, 168, 180, 182 and 184) of the derivatized (N-TFA, O-isopropyl) HCl-hydrolysed hot-water extracts of each MDRS desert soil sample and serpentine blank. The identification of the peaks is given in Table 2. The external standard (pyrene) is outside this region (retention time 90 min, m/z 202) and does not co-elute with any amino acid. X is an unidentified compound.
Table 2. Amino acid peak identification in the GC-MS chromatograms of the standards, serpentine blank and Mars analogue desert soils. Molecular ions used for identification and quantification (first ion) are also displayed. Single ion m/z 202 was selected to identify and quantify pyrene (used as an external standard)
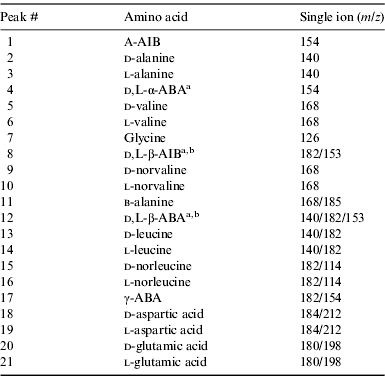
a Optically pure standard not available for enantiomeric identification.
b Enantiomers could not be separated under the chromatographic conditions.
Results and discussion
We analysed the amino acid content of several soil samples collected near the MDRS. Fig. 2 shows selected ion GC-MS chromatograms of the acid-hydrolysed hot-water extracts of the ten desert soil samples (P1–P14) and a serpentinite blank. The chromatograms were scaled to aid comparisons, with the scale factors shown in brackets. The corresponding amino acid abundances are displayed in Table 3. The most abundant amino acids were l-glutamic acid, d-glutamic acid, l-aspartic acid, l-valine, l-alanine, l-leucine and glycine (Table 3 and Fig. 2). These amino acids were also the most abundant in other hot desert soils, such as the Oman (Martins et al. Reference Martins, Hofmann, Gnos, Greenwood, Verchovsky, Franchi, Jull, Botta, Glavin and Dworkin2007), Arequipa (top of slope sample) and Atacama (flat top hill sample) desert soil samples (Peeters et al. Reference Peeters, Quinn, Martins, Sephton, Becker, van Loosdrecht, Brucato, Grunthaner and Ehrenfreund2009). Peeters et al. (Reference Peeters, Quinn, Martins, Sephton, Becker, van Loosdrecht, Brucato, Grunthaner and Ehrenfreund2009) also detected β-alanine and γ-ABA in the Atacama and Arequipa samples, which we did not detect in the MDRS soil samples.
Table 3. Summary of the average total amino acid abundances (in ppb) in several Mars soil analogues collected in the Utah desert and measured by GC-MSa

a Quantification of the amino acids included background-level correction using a serpentine blank. The associated errors are based on the standard deviation of the average value between six separate measurements (N) with a standard error, δx=σx. N −1/2.
b Enantiomers could not be separated under the chromatographic conditions.
c Optically pure standard not available for enantiomeric identification.
d Total amino acid concentrations rounded to two significant digits.
n.d., not detected.
The total amino acid abundances were very heterogeneous in the ten MDRS samples, with values ranging from no amino acids detected in soil samples P2, P5 and P6, to 100 000 ppb in sample P13 (Fig. 2 and Table 3). Sample P1 had total amino acid abundances of a few hundred ppb, samples P3, P8 and P10 contained a few thousand ppb, and samples P14 and P13 were the most abundant samples containing 17 000 and 100 000 ppb, respectively. When comparing the amino acid distribution and abundances, it is important to consider the different locations from which the soils were collected. Soil samples P5 and P6 were collected from the same location (same coordinates), samples P13 and P14 were from the same location but different depths (P13 is surface sample, while P14 is from 15 cm below the surface), and samples P8 and P10 were from the same area but with different coordinates (Fig. 1 and Table 1). P2 was collected from the same area as P3, but their total amino acid abundances are different (no amino acids were detected in sample P2). The differences in abundances between the ten soil samples may be explained by different mineral compositions, which can influence the content and stability of organic compounds in the soil (Peeters et al. Reference Peeters, Quinn, Martins, Sephton, Becker, van Loosdrecht, Brucato, Grunthaner and Ehrenfreund2009).
Analyses show that samples P5 and P6 have 23 and 33% total clay, respectively, and no detectable amino acids, while samples P13 and P14 (which are the most amino-acid-rich MDRS soil samples) have 9 and 1% total clay, respectively (Kotler et al. Reference Kotler2011). Therefore, our data indicate that soils with the higher percentage of clay are those with less extractable amino acids. Clays are expandable layers that may hold organic compounds and should protect these organic molecules against destruction by radiation and oxidizing conditions (as no radiation or oxygen penetrates the clay layers). However, once the clay layers are compacted very tightly together, organic compounds are adsorbed in the clay and it is not possible to extract the molecules. This scenario is also applicable to shales (i.e. clay-rich sedimentary rocks).
Samples P8 and P10, both from the Mancos Shale Formation, contain 4 and 5% organic matter, respectively, which are the MDRS samples that contain the highest percentage of organic matter (Orzechowska et al. Reference Orzechowska, Kidd, Foing, Kanik, Stoker and Ehrenfreund2011). Total amino acid abundances of 4600 and 2300 ppb, respectively, indicate that amino acids are also efficiently retained in these soil types or the higher organic content is due to the presence of more refractory aromatic components such as kerogen-type material. The concentration of naphthalene in samples P8 and P10 was measured to be 20 and 10 ng g−1, respectively (Orzechowska et al. Reference Orzechowska, Kidd, Foing, Kanik, Stoker and Ehrenfreund2011).
Although the clay content is important, other minerals should also be taken into account. Our data show that samples containing a high percentage of gypsum (calcium sulfate dehydrate), such as P14, P10 and P8 (Kotler et al. Reference Kotler2011), also have a high/medium amino acid content. This is in agreement with the results from Aubrey et al. (Reference Aubrey, Cleaves, Chalmers, Skelley, Mathies, Grunthaner, Ehrenfreund and Bada2006), who determined that amino acids can be preserved for geologically long periods in sulfate mineral matrices.
Differences in the total amino acid content in MDRS soils may also reflect the proliferation of any microbial population present. Notably, the total amino acid content follows the sequence Dakota Formation>Mancos Shale>Morrison Formation, while porosity and permeability follow the same trend. Higher permeability represents opportunities for microbial populations to access water, nitrogen and oxygen that can add to the organic matter present in the rock samples and therefore promote growth. Samples P7, P8, P10, P13 and P14 were collected after 23 February 2009 when 1 mm of rain was recorded (Foing et al. Reference Foing, Stoker, Zavaleta, Ehrenfreund, Direito, Kotler, Martins, Orzechowska and Thiel2011). Except for samples P6, P7 and P14, at least one (or more) domains of life were present in the soil samples (Direito et al. Reference Direito, Ehrenfreund, Marees, Staats, Foing and Röling2011; Thiel et al. Reference Foing, Stoker, Zavaleta, Ehrenfreund, Direito, Kotler, Martins, Orzechowska and Thiel2011). While we did not detect any amino acids in soil samples P6 and P7, sample P14 had a total amino acid abundance of 17 000 ppb (Table 3). Two possibilities exist to explain the differences between the amino acid and DNA data of sample soil P14: (i) The amino acids are artefacts introduced through sample handling. Further insight is provided by examining the racemic nature of the amino acids in soil sample P14. The sample contains exclusively l-amino acids reflecting the contribution of recent life. All other samples that have amino acids, and for which at least one domain of life was detected, contain a mixture of d- and l-amino acids (Table 3), indicating that racemization of the amino acids could occur over an extended period of time. (ii) Alternatively, the fact that no domain of life was detected in sample P14 is explained by zero-percent DNA recovery from a spiked P14 sample, indicating that even when a (spiked) micro-organism is present in the soil, DNA is not recovered and detected, giving a false negative (Direito et al. Reference Direito, Ehrenfreund, Marees, Staats, Foing and Röling2011).
It is also interesting to note that although bacteria were detected in soil samples P2 and P5 (Direito et al. Reference Direito, Ehrenfreund, Marees, Staats, Foing and Röling2011), we did not detect any amino acids in these soil samples (Fig. 2 and Table 3). Bacteria were detected in those two samples only with the PowerSoil kit, with no confirmation with the FastDNA kit, suggesting a false positive (Direito et al. Reference Direito, Ehrenfreund, Marees, Staats, Foing and Röling2011).
Implications for future life-detection missions to Mars
Multi-disciplinary studies of Mars analogue desert soils showing the interaction of organic, mineralogical and microbiological compositions are of extreme importance for future life-detection missions to Mars. Our results show that the mineral composition of a soil can influence the content of organic compounds. As we now have a better understanding of the mineralogical composition of Mars, as found by the Mars Exploration Rovers and Mars Express mission (Chevrier & Mathé, Reference Chevrier and Mathé2007), great care must be taken when choosing target locations for organic analysis in future life-detection missions to Mars. Samples in the MDRS area show notable variation in their amino acid contents. These variations appear to reflect the ability of certain soil types to host microbial populations and effectively preserve their organic remains. The relationship between microbial and organic analysis is another important one. While microbial analysis did not detect any micro-organisms in soil sample P14, our organic analysis detected present-life amino acid signatures. On the other hand, results show the presence of bacteria in two soil samples (P2 and P5) where no amino acid has been detected. Although Direito et al. (Reference Direito, Ehrenfreund, Marees, Staats, Foing and Röling2011) did not determine the abundance of (potential) bacteria in these two soil samples, we know that the limit of detection of our GC-MS is 10 pg of amino acid in a sample. The Viking landers also did not detect any organic compounds on Mars above the detection limit of a few ppb (Biemann et al. Reference Biemann, Oro, Toulmin, III, Orgel, Nier, Anderson, Simmonds, Flory, Diaz and Rushneck1976, Reference Biemann, Oro, Toulmin, Orgel, Nier, Anderson, Simmonds, Flory, Diaz and Rushneck1977), but the Viking Labeled Release experiment established the presence of microbial activity (Levin and Straat, Reference Levin and Straat1976). This was explained by the destruction of organic molecules due to intense UV flux on Mars and the highly oxidizing nature of the Martian surface (Benner et al. Reference Benner, Devine, Mateeva and Powell2000; Squyres et al. Reference Squyres, Grotzinger, Arvidson, Bell, Calvin, Christensen, Clark, Crisp, Farrand and Herkenhoff2004). Alternatively, it has been suggested that the Viking organic analysis instrument was not sensitive enough and could not detect the degradation products generated by several million bacterial cells per gram of Martian soil (Glavin et al. Reference Glavin, Schubert, Botta, Kminek and Bada2001). As preparation for future life-detection space missions, it is crucial to perform further parallel mineralogical, microbial and organic analyses of several Mars desert soil analogues, and determine the limit of detection of the experimental methods used.
Conclusions
We have analysed the amino acid content of several Mars analogue soil samples collected near the MDRS, in Utah (USA). The results show a large variation between the soil samples, even the ones that were collected from the same location. Three soil samples (P2, P5 and P6) did not contain any amino acid, while two other samples (P13 and P14) had a high amino acid content. Our data indicate that the mineral composition of the soil strongly influences its organic composition or the extractability of organic material from the mineral matrix. Soil samples that contained a high percentage of clay did not have any detectable amino acids. This has implications for future life-detection missions to Mars, as locations with low (or absent) clay content should be targeted if we are to detect the molecular signatures of life.
Acknowledgements
Zita Martins was supported by the Royal Society. Pascale Ehrenfreund was supported by the NASA Astrobiology Institute (NAI). Zita Martins and Mark A. Sephton acknowledge the Science and Technology Facilities Council (STFC) for financial support. The EuroGeoMars 2009 campaign was organized and supported by the International Lunar Exploration Working Group (ILEWG), NASA Ames Research Centre and ESA/ESTEC. We acknowledge the contribution of the EuroGeoMars 2009 campaign crew and the mission support team. This research was conducted in the framework of the Mars Express Recognized Cooperating Laboratory for geochemistry.