Vancomycin-resistant Enterococcus (VRE) pose a significant problem in healthcare settings across the world.Reference Satilmis, Vanhems and Benet1 Infections are often preceded by colonization, where bacteria are carried in the gastrointestinal system asymptomatically.Reference Akturk, Sutcu and Somer2–Reference Kara, Devrim and Bayram4 Studies have shown that a significant proportion of patients in the healthcare system are colonized and have an elevated risk of infection and subsequent morbidity and mortality. Although VRE may be less virulent than some other multidrug-resistant organisms, the limited treatment options are a cause for concern, particularly for highly vulnerable patient populations such as preterm neonates, for whom infections remain a significant cause of mortality.Reference Khan, Morris and Bhutta5 Many guidelines and efforts are therefore aimed not only at preventing infections but also at reducing colonization. In a hospital setting, it is assumed that VRE is transmitted between patients via the hands of healthcare staff or through the hospital environment.Reference Flokas, Karageorgos, Detsis, Alevizakos and Mylonakis3 Neonates requiring intensive care have many factors predisposing them for colonization, and VRE may quickly spread within this patient group.Reference Iosifidis, Evdoridou and Agakidou6, Reference Malik, Montecalvo and Reale7
Between January and May in 2017, the Canberra Hospital (CH) experienced its first outbreak of VRE colonization in the neonatal intensive care unit (NICU) and the special care nursery (SCN). The situation was swiftly brought under control by the rapid implementation of infection control measures that led to the termination of transmission. However, identification of the origin and transmission pathway remained to be elucidated, and these factors are the subject of this study.
Methods
Description of the outbreak and response measures
The outbreak was identified following the detection of VRE in 4 neonates transferred from CH to a secondary-care facility in Canberra between January 27, 2017, and March 9, 2017. Active screening culture (ASC) protocols were implemented at CH on March 15, 2017, which encompassed screening of neonates at first bowel movement after admission, followed by weekly screening. Fecal samples were collected on swabs and cultured directly onto chromogenic chromID VRE agar plates (bioMèrieux, Marcy-l’Étoile, France) and incubated for at least 48 hours. Enterococcus spp were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF MS; Bruker Daltronics, Billerica, MA) and vancomycin resistance was confirmed by Vitek 2 (bioMerieux). Van genotype was determined by Xpert vanA/vanB (Cepheid, Sunnyvale, CA) polymerase chain reaction (PCR). Screening identified 8 patients in the NICU and SCN to be colonized with VRE. During the following ASC period another 2 patients were found to be colonized with VRE. The outbreak was declared contained on the May 26, following 3 weeks (24 days) without a positive screening culture. Standard personal protective equipment for MRO events with full gown and gloves were used by the staff during the outbreak period. It is possible that additional infants were colonized who were discharged prior to the commencement of the ASC.
In addition to the ASC protocols, contact precautions for all VRE-colonized patients was implemented and cohort isolation of VRE-colonized patients into specific areas of the 2 wards. A VRE working group was established, and review of infection control policies, operational protocols, and cleaning practice in the 2 units was also performed. Microbial testing of high-touch environmental surfaces and equipment commonly used between infants was conducted, including weight scales, length measurer, IV trolleys, ultrasound equipment, cord of the blinds, exit door plate to NICU and SCN, bottle warmers, and door handles to isolation rooms. Moistened BD CultureSwab (Becton Dickinson, Franklin Lakes, NJ) were used for environmental sampling and were sent in Stuart’s transport media for microbiological analysis. None of the environmental samples tested positive during the ASC screening period.
Setting and population
The Canberra Hospital is an adult and pediatric tertiary-care center with 672 beds serving a population of ~550,000. The neonatal care service is composed of 2 sections: the NICU with 21 beds in 2 bed bays with 1 isolation room and the SCN with 15 beds in 2 bed bays with 1 isolation room. The NICU provides intensive care for seriously ill or premature infants or infants requiring a high level of observation. The SCN provides step-down care for infants from the NICU or those infants directly admitted requiring care outside the NICU.
Study design
A retrospective case control study was conducted, capturing all patients admitted in the NICU and SCN during the period from December 18, 2016, to May 26, 2017. The time frame covers the outbreak period and was defined by the earliest admission date of a VRE-positive patient and the latest discharge date of a VRE-positive patient.
Variables and definitions
Colonization was used as the outcome variable. Cases were defined as patients admitted to the NICU and SCN during the period from December 18, 2016, to May 6, 2017, and had 1 or more cultured specimen positive for a VRE. Controls were defined as any patient admitted to the NICU or SCN during the same period and had no positive cultured specimens and at least 1 negative cultured specimen for a VRE.
Plausible exposure variables were identified from published literature and from discussions with infectious disease and infection prevention and control specialists. Patient variables collected are listed in Table 1.
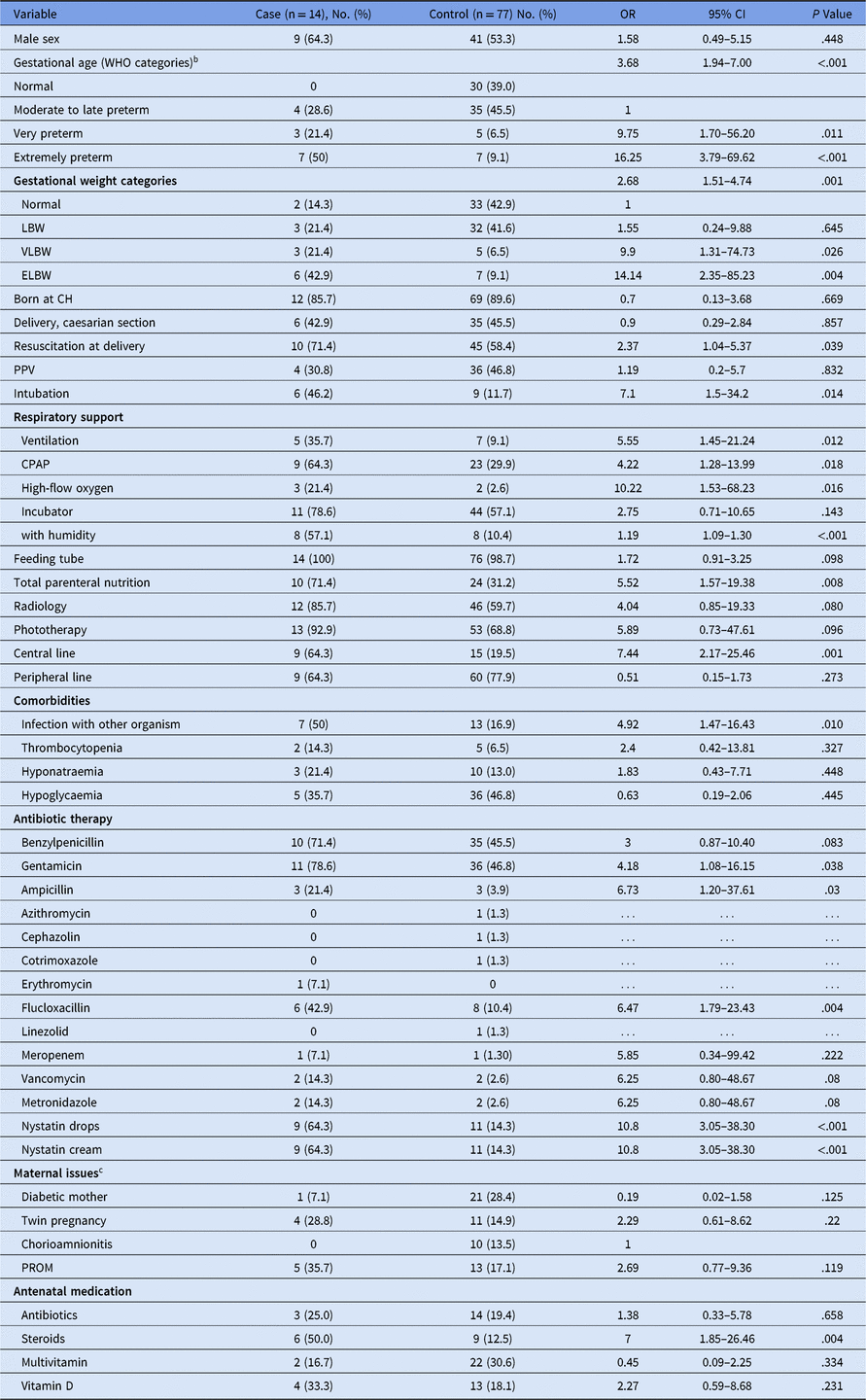
Table 1. Univariate Logistic Regression Analysis of Risk Factors for Colonization With Vancomycin-Resistant Enterococcus (VRE)a

Note. OR, odds ratio; CI, confidence interval; WHO, World Health Organization; LBW, low birth weight; VLBW, very low birth weight; ELBW, extremely low birth weight; TCH, Canberra Hospital; PPV, positive predictive value; CPAP, continuous positive airway pressure; PROM, prelabor rupture of membranes.
a Variables are given as no. (%) unless otherwise stated in the variable title. For continuous variables, the mean ± standard deviation (SD) is given.
b For factor-variable univariate analysis the “normal” and “late to moderate” categories were combined and used as the reference category.
c For 3 patients the maternal data were not available.
Data analysis was carried out using Stata version 15 software (StataCorp, College Station, TX). To determine significant risk factors for colonization, univariate logistic regression was conducted on all variables. Variables with a P < .10 in the univariate analysis were selected for inclusion in a backward-stepwise multivariate logistic regression model with the P-value threshold for elimination set at .05. The relationship between variables was assessed using Pearson’s correlation coefficients and collinearity analysis.
Whole-genome sequencing
In total, 11 VRE isolates obtained from patients in the outbreak were referred to the Microbiological Diagnostic Laboratory Public Health Laboratory for whole-genome sequencing (WGS). Isolates were not available from the first 3 patients in the outbreak. Genomic DNA was extracted using the JANUS automated workstation with the Chemagic Viral DNA/RNA kit (PerkinElmer, Waltham, MA). Unique dual indexed libraries were prepared using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA). Libraries were sequenced on the Illumina NextSeq 500 with 150-cycle paired end chemistry as described by the manufacturer’s protocols. The Nullarbor pipeline (version 2.0, https://github.com/tseemann/nullarbor) was used for all bioinformatics analyses with an in-house ST1421 reference genome. Gubbins was used to detect regions of recombination that were removed, and IQtree was used to infer the maximum likelihood phylogeny from the resulting core SNP alignment.Reference Croucher, Page and Connor8–Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel11
Ethics
Ethics approval for the study was obtained from the ACT Health Human Research Ethics Committee (Protocol number ETHLR.17.155) and from the Australian National University Human Research Ethics Committee (Protocol number 2017/536).
Results
Outbreak cohort
During the study period from December 18, 2016, to May 6, 2017, 91 patients were admitted to the NICU and SCN at Canberra Hospital (TCH). Of these, 14 were colonized with VRE during their admission. The remaining 77 were negative for VRE throughout their admission, resulting in a VRE prevalence of 15.4%.
Risk factor analysis
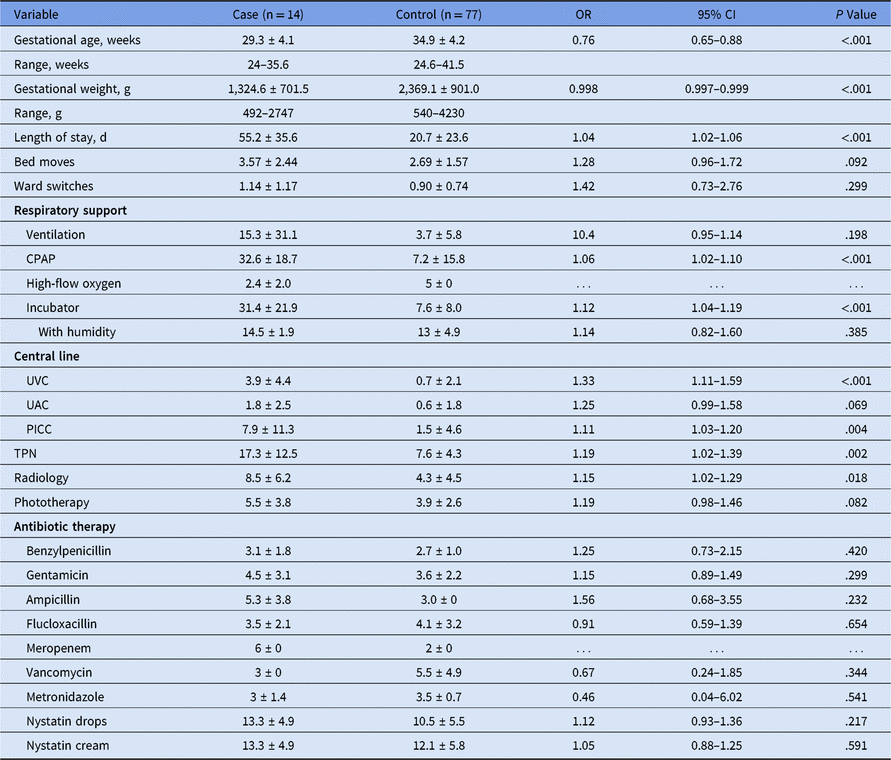
Univariate logistic regression was performed to assess the predictive value of individual variables on the colonization outcome. Increased odds of colonization were observed for early gestational age, low birth weight, length of stay, resuscitation at delivery, requirement for respiratory support with ventilation or continuous positive airway pressure (CPAP) or high-flow oxygen, use of incubator with humidity, infection with other organisms, total parenteral nutrition (TPN), radiology, phototherapy, presence of central lines, select antibiotics, and antenatal steroids (Table 1). Significant differences in duration of use were seen for CPAP, incubator use, central lines, TPN, and radiology (Table 2). Interestingly, the mean durations of antibiotics use were not significantly different between colonized and noncolonized patients.
Table 2. Risk Factor Variables for Colonization with Vancomycin-Resistant Enterococcus (VRE) with Continuous Scalesa

Note. OR, odds ratio; CI, confidence interval; UVC, Umbilical vein catheter; UAC, Umbilical artery catheter; PICC, peripherally inserted central catheter; TPN, total parenteral nutrition.
a Stated as (mean ± standard deviation) duration in days unless otherwise indicated in variable title. The mean does not include patients for which the variable was not used.
Variables with a P < .10 in the univariate analysis were selected for inclusion in a backward stepwise multivariate logistic regression model. Collinearity was observed with the benzylpenicillin and gentamicin variables, and with nystatin cream and nystatin drops, where inclusion of both in the multivariate model resulted in a failure to converge. The final reduced multivariate model showed significant association with only gestational age (WHO preterm categories), with an odds ratio (OR) of 3.68, (95% CI, 1.94–7.00).
Whole-genome sequencing
Vancomycin-resistant E. faecium isolates from 11 of the infants in the outbreak were underwent WGS. In silico multilocus sequence typing (MLST) showed that all the isolates were sequence type 1421 (ST1421), with the allelic profile atpA(1), ddl(1), gdh(1), purK(1), gyd(1), pstS(0), adk(1). These pstS null strains were previously classified as nontypeable but have now been assigned sequence types in the MLST scheme, following the definition of pstS null alleles as 0. ST1421 is a single-locus variant of ST17. The genomic analysis confirmed the initial PCR-based results showing the presence of the vanA gene.
Phylogenetic reconstruction of the 11 isolates are shown in context with 7 E. faecium ST1421 vanA isolates identified from the orthopedic ward of CH collected over a 9-day period in late 2016 and 27 consecutive E. faecium ST1421 vanA isolated during a 1-month period in late 2015 at the ACT Pathology (including TCH and other ACT hospitals) (Fig. 1A). The violin plot shows that all 11 isolates were highly clonal with single nucleotide polymorphism (SNP) distances ranging from 0 to 5 core genome substitutions (Fig. 1B), a considerably smaller range compared with that of the sample set as a whole (0–59 SNPs). The orthopedic isolates which were collected over a 9-day period show a much greater pairwise SNP range (2-44 SNPs), and with 3 separate clusters in the phylogenetic tree. Sequence reads are available in the Sequence Read Archive at the National Center for Biotechnology Information, under BioProject PRJNA495857 (Supplementary Table 1 online).

Fig. 1. Whole-genome sequencing data for isolates from 11 of the infants in the outbreak (isolates not available from 3 infants), 7 ST1421 isolates from the orthopedic unit at TCH and 27 ST1421 submitted from ACT Pathology during the 2015–2016 Victorian snapshot. (A) Maximum likelihood phylogenetic reconstruction based on core-genome SNPs. The neonatal intensive care unit (NICU) outbreak cluster is highlighted in pink and orthopedic clusters are highlighted in teal. (B) Violin plots showing the range of pairwise SNP distances between isolates within each of the cohorts, NICU outbreak isolates (pink), orthopedic isolates (teal), and other ACT Pathology isolates (purple), and the intercohort (grey). The mean, median, and interquartile ranges of pairwise SNP distances ae shown.
Patient ward and bed movements
Colonized patients had an average of 3.6 bed movements during their admission, and noncolonized patients had an average of 2.7 bed movements, the difference being insignificant. Even less difference was seen in movement between the NICU and SCN wards, with averages of 1.14 and 0.90 for colonized and noncolonized patients, respectively. In some instances, patients were moved into bed spots with time gaps as short as 29 minutes and 60 minutes following the previous patient.
Discussion
The outbreak subject to this investigation represents the first known occurrence of VRE colonization in the NICU and SCN wards at CH. Neonates in these units have no contact with other patients and interaction with a limited number of healthcare workers, immediate caregivers, and the hospital environment. VRE colonization in this patient group must therefore be a direct result of a breakdown in infection control procedures within these units. Colonization and infection with VRE have been identified among patients in other wards of CH, suggesting that a cross-contamination event could have led to the introduction of VRE into the NICU/SCN environment, similar to that described in a previous study.Reference Pusch, Kemp and Trevino13 Once the environment had been contaminated, it is possible that procedures within the units led to dissemination of VRE to several patients.
The range of variables found to be associated with colonization at a univariate level in our study is consistent with those observed in the literature. The range of variables associated with VRE colonization previously observed in NICU outbreaks include: prematurityReference Malik, Montecalvo and Reale7, Reference Pusch, Kemp and Trevino13–Reference Miedema, Kerkhof, Arends, Bergman and Kimpen15; low birth weightReference Malik, Montecalvo and Reale7, Reference Pusch, Kemp and Trevino13–Reference Tanyeri-Bayraktar and Bayraktar16; prepartal antibioticsReference Pusch, Kemp and Trevino13; longer duration of antibiotic therapyReference Malik, Montecalvo and Reale7, Reference Hufnagel, Liese and Loescher14, Reference Tanyeri-Bayraktar and Bayraktar16, Reference Yuce, Karaman, Gulay and Yulug17; administration of second-line antibioticsReference Iosifidis, Evdoridou and Agakidou6; long hospitalisationReference Malik, Montecalvo and Reale7, Reference Hufnagel, Liese and Loescher14, Reference Miedema, Kerkhof, Arends, Bergman and Kimpen15, Reference Yuce, Karaman, Gulay and Yulug17, Reference Devrim, Genel, Atlihan, Özbek and Gülfidan18; mechanical ventilationReference Hufnagel, Liese and Loescher14, Reference Miedema, Kerkhof, Arends, Bergman and Kimpen15; presence of a central lineReference Malik, Montecalvo and Reale7, Reference Hufnagel, Liese and Loescher14, Reference Miedema, Kerkhof, Arends, Bergman and Kimpen15; presence of peripheral linesReference Malik, Montecalvo and Reale7; parenteral nutritionReference Malik, Montecalvo and Reale7, Reference Hufnagel, Liese and Loescher14, Reference Miedema, Kerkhof, Arends, Bergman and Kimpen15; major surgeryReference Hufnagel, Liese and Loescher14; episodes of clinical features compatible with infectionReference Hufnagel, Liese and Loescher14; nasogastric feedingReference Malik, Montecalvo and Reale7, Reference Miedema, Kerkhof, Arends, Bergman and Kimpen15; ultrasonographyReference Miedema, Kerkhof, Arends, Bergman and Kimpen15; administration of CPAPReference Malik, Montecalvo and Reale7; use of oxyhoodReference Malik, Montecalvo and Reale7; procedures by non-NICU staffReference Malik, Montecalvo and Reale7; consultations by non-NICU staff.Reference Malik, Montecalvo and Reale7 Many of these risk factors are linked with each other, and they likely are indicators for a common risk factor.
In this study, after adjusting for all other variables, only gestational age remained significantly associated with colonization. Our results agree with published data. In previous studies that have conducted a multivariate analysis, most observed a similar drastic reduction in the number of associated variables: gestational age onlyReference Pusch, Kemp and Trevino13; gestational age, longer duration in hospital, use of antibiotics and presence of central linesReference Hufnagel, Liese and Loescher14; birth weight and days of antimicrobial therapyReference Malik, Montecalvo and Reale7; administration of second-line antibiotic and hospital admission during specific periodReference Iosifidis, Evdoridou and Agakidou6; gestational age; anti-MRSA therapy; and ultrasonography.Reference Miedema, Kerkhof, Arends, Bergman and Kimpen15 There may be several mechanisms by which gestational age contributes to colonization. Premature infants have been shown to become colonized with a lower species complexity, composed of few Bifidobacteria spp, and higher rate of potentially pathogenic bacteria.Reference Khan, Sarwari and Hasan19–Reference Jacquot, Neveu and Aujoulat23 The establishment of a gut microbiome is further disrupted by empiric use of antibiotics.Reference Drell, Lutsar and Stsepetova20, Reference Gewolb, Schwalbe, Taciak, Harrison and Panigrahi22, Reference Patel, Mutlu and Sun24 Furthermore, birth at an early gestational age usually results in a longer hospital stay, thereby increasing the exposure time to potentially contaminated healthcare settings.
As there was no routine testing for VRE conducted for admitted patients prior to the outbreak response, it is difficult to establish with certainty when the introduction of VRE into the units occurred. The frequent transfers to the secondary-care hospital, which routinely test transferred infants for MROs, suggests that this was a relatively recent introduction because there had not been any previous alerts. As most of the patients were admitted prior to the ASC screening, they may have been colonized at any previous time point. Two infants may have had exposures outside TCH. One 1 infant was admitted on day 41 of life, and the other was born at TCH but was transferred to another hospital for a procedure after which the patient was readmitted to the NICU at TCH. Two patients were admitted after the commencement of the ASC screening. Their admission histories show only exposure to the SCN ward, while some other patients only had exposure to the NICU ward. Thus, both wards were likely to be contaminated and had transmission events.
Following the alert of VRE-positive infants transferred to the secondary-level hospital, a bundle of measures was implemented to control the outbreak. Several of these measures have previously proven successful in the eradication of large-scale VRE outbreaks, such as formation of a VRE executive working group, mass screening, cohort isolation of carriers, environmental screening, and review of cleaning procedures and operational protocols.Reference Madan, Salari and Saxena25 It is well recognized that the hospital environment can be a reservoir for VRE.Reference Flokas, Karageorgos, Detsis, Alevizakos and Mylonakis3, Reference Christiansen, Tibbett and Beresford26–Reference Kramer, Schwebke and Kampf28 In our study there were no positive swabs indicating environmental contamination, although this could also be due to the failure to identify the contaminated sites or to sample or methodological failures, such as inhibition due to residual cleaning agents on the sampled surfaces. The units are part of the national hand hygiene program, and they passed an audit in early 2017. The frequent bed movement of patients is a potential concern, especially if there is insufficient cleaning of the evacuated bed spot between patients. Hospital protocol states a minimum standard gap of 45 minutes, extended to 90 minutes for colonized patients. Following the outbreak, the bed-cleaning protocol as well as the NICU policy to only conduct bed moves on clinical grounds were reinforced.
A genomic analysis of the rise of Australian E. faecium vanA in recent years showed that the expansion was polyclonal and not the result of expansion of a single successful clone.Reference Otter, Yezli, Salkeld and French29 The 2014 Australian Enterococcal Sepsis Outcome Program (AESOP) showed that E. faecium had 5 dominating sequence types: ST17, ST117, ST203, ST555, and ST796.Reference van Hal, Espedido and Coombs30 An interesting feature in Australia is the recent emergence of healthcare-associated E. faecium strains where the pstS allele has been deleted from the genome, described by Carter et al. Reference Coombs, Daley and Thin Lee31 These strains were present in multiple Australian jurisdictions and nearly universally displayed a vancomycin-resistant phenotype. There was significant diversity in the core genome among the 66 studied isolates, with a maximum pairwise SNP distance of 4,543. The phylogenetic reconstruction showed clustering of isolates by hospitals. In a recent study of 240 E. faecium vanA isolates from 12 ICUs in New South Wales, Australia, 60.0% were ST1421 and 24.2% were ST1424 (allelic profile atpA(9), ddl(1), gdh(1), purK(1), gyd(12), pstS(0), adk(1)).
The range of pairwise SNP distances within the 45 samples analyzed in the present study is considerably smaller than that described for the national sample set presented by Carter et al, Reference Coombs, Daley and Thin Lee31 supporting the observation of clustering of clones by hospitals described in Carter et al. and van Hal et al. Reference Otter, Yezli, Salkeld and French29 The ST1421 samples from the NICU outbreak form a tight cluster clearly derived from a ST1421 clonal lineage endemic in the CH environment. The samples were highly clonal, with short internal branch lengths suggestive of a recent point source introduction and subsequent transmission within the unit. In contrast, the dispersed nature of the samples from the orthopedic unit distributed between multiple clusters in the phylogenetic tree is indicative of multiple exposures and introductions, which is interesting considering the relatively short time period over which these samples were collected. The orthopedic ward has a patient population with a much more diverse exposure history, compared with the naïve patient population of the NICU. The data support the hypothesis that VRE causing colonization in the NICU/SCN was likely acquired from cross contamination in the hospital, rather than being imported from the community. We were unable to screen staff or care givers; notably, however, anecdotal evidence indicated that some staff worked also in other hospital wards.
This study has 2 main limitations. First, the lack of routine VRE screening makes it difficult to determine the exact time point of VRE acquisition in patients and the introduction of the organism into the units. Second, the dearth of data on VRE in the wider community outside the healthcare system limits our ability to exclude the possibility that the VRE observed in the NICU and SCN was introduced from a community source, rather than from an intrahospital transmission event.
The presence of a disseminated VRE clone within the hospital increases the probability of future outbreaks. Biannual screening of the NICU has now been implemented, which will lead to earlier detection of colonization, as has previously been described in the literature.Reference Carter, Buultjens and Ballard32 It is encouraging that no new risk factors for colonization other than those previously described in the literature were identified in this study and that the outbreak was contained relatively easy, meaning that transmission can be controlled even in a largely endemic environment.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2019.41
Author ORCIDs
Patiyan Andersson, 0000-0003-2705-1847
Acknowledgments
We would like to acknowledge the effective response of the staff in the Neonatal Intensive Care Unit, the Special Care Nursery and the Infection Prevention and Control Unit at the Canberra Hospital in containing this outbreak and for their assistance in this investigation. We also would like to thank ACT Pathology for their assistance in providing isolates for sequencing.
Conflicts of interest
P. Andersson is supported by a 2-year scholarship from the Australian Government Department of Health during his field epidemiology training program, Master of Philosophy (Applied Epidemiology). All authors report no conflicts of interest relevant to this article.