Carbapenem-resistant Enterobacteriaceae has spread across the United States and worldwide in the decade.Reference Munoz-Price, Poirel and Bonomo 1 The Centers for Disease Control and Prevention recently categorized carbapenem-resistant Enterobacteriaceae as one of the organisms posing an urgent threat to public health. Among the several mechanisms underlying resistance to carbapenems in the family Enterobacteriaceae, carbapenemase production is of particular interest for healthcare epidemiology because of the ability of plasmids encoding carbapenem-resistant Enterobacteriaceae genes to transmit between species.Reference Nordmann and Poirel 2 Among carbapenemase-producing Enterobacteriaceae (CPE) in the United States, Klebsiella pneumoniae carbapenemase (KPC) is the most frequently identified carbapenemase.Reference Munoz-Price, Poirel and Bonomo 1 The KPC gene, bla KPC, is encoded on transferable, promiscuous plasmids in most instances, allowing it to spread to species beyond K. pneumoniae.Reference Mathers, Cox and Kitchel 3 Cases of KPC-producing Enterobacter spp. and Escherichia coli have been reported.Reference Ahn, Syed, Hu, O’Hara, Rivera and Doi 4 , Reference O’Hara, Hu and Ahn 5 Citrobacter freundii belongs to the family Enterobacteriaceae and is a well-recognized, if not frequent, organism responsible for healthcare-associated infections.Reference Zarb, Coignard and Griskeviciene 6 KPC-producing C. freundii cases have been reported, but they remain sporadic.Reference Adler, Khabra, Paikin and Carmeli 7 , Reference Tijet, Muller, Matukas, Khan, Patel and Melano 8
In March 2015, the infection control department at a major teaching hospital in Florida identified an increase in the number of carbapenem-resistant C. freundii isolates reported by the clinical microbiology laboratory. This report aims to describe the investigation and control of this unusual outbreak of carbapenem-resistant C. freundii that produced KPC.
METHODS
Study Setting
This outbreak investigation took place at a 1,500-bed acute care facility in Miami, Florida, from December 1, 2014, through June 30, 2015. The hospital is a referral national and international center offering services in a wide range of specialties, including a large transplant institute and the only level 1 trauma center in the region. The facility has previous experience with patients colonized or infected by CPE.Reference Munoz-Price, De La Cuesta and Adams 9 Historically, most CPE-colonized or CPE-infected patients are identified in the surgical intensive care unit (SICU) and the transplant units. The SICU is physically divided into 2 areas, SICU-A and SICU-B; SICU-A is an open-bay unit with 20 beds and SICU-B offers 20 private beds.
Case Definition
Cases were defined as any patient with C. freundii isolated from either a surveillance or a clinical culture and reported as resistant to any of the carbapenems by the antimicrobial susceptibility testing using Clinical and Laboratory Standards Institute breakpoints guidelines 10 in the Vitek 2 system (bioMérieux) and that also were positive for carbapenemase production by the Carba NP test (bioMérieux).Reference Nordmann, Poirel and Dortet 11 Surveillance cultures were performed by obtaining a perirectal swab sample that was subsequently cultured on a MacConkey agar plate with an ertapenem disk. We conducted a retrospective search for patients who met case definition by reviewing the Infection Control Department’s automatic surveillance system (Vigilanz) from January 1, 2014, through July 1, 2015; only incident cases were included. As part of the routine operations of the department, the surveillance system alerts the infection preventionists in real time with any gram-negative bacilli reported as carbapenem-resistant, triggering contact isolation as part of the standard infection control practice of the hospital.
Data were collected using the electronic medical record and included patient demographic characteristics, comorbidities, transfer history within the facility, and use of invasive devices.
Microbiology and Molecular Typing
Further characterization of the isolates involved in the outbreak was conducted at the University of Pittsburgh. The presence of bla KPC in the C. freundii isolates was identified by polymerase chain reaction.Reference O’Hara, Hu and Ahn 5 Minimum inhibitory concentrations (MICs) of an array of antimicrobial agents including carbapenems were obtained by the broth microdilution method using commercially available plates (Sensititre GNX2F; Thermo), except for carbapenems (ertapenem, imipenem, and meropenem), which were tested with Etest (bioMérieux) to obtain more granular MICs. Relatedness of the C. freundii isolates was initially determined by pulsed-field gel electrophoresis using XbaI as the restriction enzyme as previously reportedReference Alrowais, McElheny and Spychala 12 and interpreted visually according to the Tenover criteria.Reference Tenover, Arbeit and Goering 13
To define the sequence types, resistome, and whole genome phylogenetic relationship of the isolates, the genomes were extracted by DNeasy Blood and Tissue Kit (Qiagen) for library construction and sequenced on NextSeq (Illumina) using the paired-end mode. De novo assembly of the sequence reads was performed using CLG Genomics Workbench 7 (Qiagen). Multilocus sequence typing was conducted in silico from the contigs. New alleles were confirmed with polymerase chain reaction and Sanger sequencing using the recommended protocol (http://pubmlst.org/cfreundii/) and registered. Antimicrobial resistance genes were identified using ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) and a cutoff of 80% similarity. Whole genome phylogenetic relationships among the 5 KPC-producing C. freundii genomes in this study were compared with other publicly available C. freundii genomes on Genbank. An alignment of 2,602 shared genes (core genome) was obtained using RoaryReference Page, Cummins and Hunt 14 and a maximum likelihood phylogenetic tree with 1,000 rapid bootstrap replicates was generated using RAxML.Reference Stamatakis 15 Additionally, pairwise single-nucleotide polymorphism differences between 5 KPC-producing C. freundii genomes in this study were generated using Nullarbor (https://github.com/tseemann/nullarbor) with isolate M1 as an internal reference and extracted using a custom analytic pipeline.
Bundle of Interventions
As previously reported, the facility implemented a series of interventions to prevent transmission of CPE in 2009.Reference Munoz-Price, De La Cuesta and Adams 9 The bundle of interventions in place in 2014, at the moment of this outbreak, included the following components: active surveillance cultures in the adult intensive care units to identify carriage of CPE using perirectal swab (and also using tracheal aspirate if the patient was mechanically ventilated), on admission and weekly thereafter; surveillance cultures were processed placing a 10-μg meropenem disk in the main streak of a blood agar plate and a MacConkey plate with further identification and Carba NP testing of those organisms that grew near the meropenem disk. The infection control bundle included, in addition to the surveillance cultures, daily bathing with 2% chlorhexidine-impregnated wipes (Sage) of all adult patients admitted in the intensive care units or transplant units; use of bleach wipes for daily disinfection and terminal cleaning in the intensive care units by environmental services and quaternary ammonium (Super Sani Cloth; PDI Healthcare) for per-shift disinfection of surfaces considered to be close to the patient by nursing; contact precautions and private room setting for all known CPE patients until hospital discharge and during further readmissions; staff cohorting or reduced nurse/patient ratio for the nurses taking care of CPE patients; use of disposable blood pressure cuffs, stethoscopes, and thermometers and use of other dedicated equipment; change of disposable curtains after terminal cleaning in all intensive care unit rooms and all rooms occupied by patients in contact precautions; labeling of the electronic medical record to facilitate identification of CPE patients during transfers or readmissions; daily emails to the nursing administration and bed placement service with the list of CPE patients admitted in the facility to facilitate staff cohorting started in August 2014.
In response to the outbreak described here, supplementary environmental cleaning was implemented in addition to the interventions already in place at the time. In the first week of April 2015, all CPE patients present in SICU-B, where most cases occurred, were moved to a new pod of terminally cleaned private rooms within SICU-B, followed by closure and terminal cleaning of the pod where the patients were located before, and a special request was made to the environmental services to provide extra personnel and time for deep cleaning of the affected rooms and surrounding area. Around this time, the unit went into plumbing repairs related to blockage of the dialysis hookup in some rooms; owing to the blockage the dialysis effluent was drained into the hand washing sinks, making them unavailable for hand washing. This blockage was identified in early March and several rooms in SICU-B were affected. The plumbing repair took effect in April following the movement of the patients. The staff received additional education on the importance of hand hygiene and contact precaution compliance to prevent transmission of CPE the first week of May.
Those patients outside SICU-B identified as carriers of CPE were placed in contact precautions in private rooms with 1:1 nurse/patient ratio during their stay in the hospital in addition to the bundle of interventions already in place.
RESULTS
A total of 6 patients with C. freundii resistant to any of the carbapenems and positive for carbapenemase production by the Carba NP test were identified through the Infection Control Department database search. Four patients were male and 2 were female (Table 1). Median total length of stay was 108 days and median length of stay to the positive cultures was 57 days. One patient expired, resulting in a crude in-hospital mortality of 17%; the reason for this death was not related to CPE infection. Four of the patients developed carbapenemase-producing C. freundii infections; 3 of these patients were treated empirically with cefepime and 1 with meropenem and each was later switched to amikacin, levofloxacin, or trimethoprim-sulfamethoxazole for definitive therapy. All patients had surgery during their admission, but there was no association with the time or place of the procedures among the cases.
TABLE 1 Characteristics of 6 Patients Affected by the Carbapenemase-Producing Citrobacter freundii Outbreak
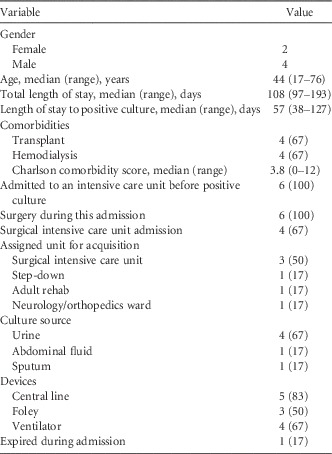
NOTE. Data are no. (%) of patients unless otherwise indicated.
All patients had rectal screening cultures and 1 patient also had tracheal aspirate screening culture collected before clinical cultures identified carbapenemase-producing C. freundii, with a range from 1 to 11 surveillance cultures per patient. The median time from the last negative surveillance culture to identification of carbapenemase-producing C. freundii in a clinical culture was 7 days. In addition, 4 patients had from 1 to 5 perirectal surveillance cultures collected after carbapenemase-producing C. freundii was identified in clinical culture. However, none of the surveillance cultures was positive for the organism (Table 2). The sources of the positive clinical cultures were urine (4), abdominal fluid (1), and sputum (1). Two of the patients had carbapenemase-producing C. freundii isolated from more than 1 body site. At the time, all 6 isolates were resistant to cefotaxime, ceftazidime, ertapenem, and meropenem and tested positive for carbapenemase production by the Carba NP test. They were variably resistant to aminoglycosides, fluoroquinolones, and tetracyclines.
TABLE 2 Surveillance Cultures Collected From Patients With Carbapenemase-Producing Citrobacter freundii

NOTE. CP, carbapenemase-producing; SC, surveillance cultures.
Before the onset of the outbreak, the median daily number of patients either colonized or infected with CPE present in the facility was 7 patients per day and mostly represented by K. pneumoniae, and most of them were first identified by rectal surveillance cultures. Most of these patients were transplant patients with previous stays at different healthcare facilities in the United States and abroad. Only 1 patient with carbapenemase-producing C. freundii was identified in 2014 (M6). At the moment of the C. freundii outbreak, there were 2 other patients with CPE other than C. freundii admitted to SICU-B. These 2 patients were not part of the outbreak.
A total of 6 patients with carbapenemase-producing C. freundii were identified in the period from December 1, 2014, through June 30, 2015 (Figure 1). In March 2015, patients M1, M2, and M3 were identified as having carbapenemase-producing C. freundii; these 3 patients were associated with SICU-B, and they were located in adjacent rooms at the moment of identification of carbapenemase-producing C. freundii in their clinical cultures. Patient M4 was identified in the last week of April 2015, and this acquisition occurred in one of the neurology/orthoplastics units; this patient had previously been in SICU-B. Patient M5 yielded the first carbapenemase-producing C. freundii isolate in the adult rehabilitation unit in June 2015. Patient M6 was identified retrospectively by reviewing the database and acquisition was associated with a step-down unit in December 2014. Patients M5 and M6 were never in SICU-B. All patients were admitted to one of the hospital’s intensive care units before acquisition of carbapenemase-producing C. freundii.
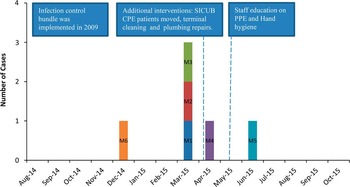
FIGURE 1 Timeline of the outbreak. A bundle of interventions was implemented in 2009. Daily emails started in August 2014. Additional interventions to control the outbreak took place between April and May 2015. CPE, carbapenemase-producing Enterobacteriaceae; PPE, personal protective equipment; SICU-B, surgical intensive care unit B.
After the third case was confirmed in SICU-B at the end of March, the unit was notified of the possible cluster with recommendations about proper hand hygiene and use of personal protective equipment. All CPE patients in SICU-B were moved to terminally cleaned rooms at the beginning of April 2015 and plumbing repairs were started also around this time. The last carbapenemase-producing C. freundii case was identified in the facility in June 2015, and no additional cases were identified up to November 2015.
Carbapenemase-producing C. freundii isolates were available from 5 of the 6 patients for further testing in the research laboratory (M1-M5). All 5 isolates carried bla KPC-3. Isolates M1-M3 were resistant to all carbapenems as well as cephalosporins, whereas isolate M4 was only intermediate to ertapenem and M5 was susceptible to all carbapenems despite resistance to 1 or more third-generation cephalosporins as well as aztreonam (Table 3). Overall, isolates M1-M3 showed comparable MIC profiles for both β-lactam and non-β-lactam agents. All isolates were susceptible to tigecycline and colistin. The susceptibility patterns generally corroborated the resistance genes identified through whole genome sequencing (Table 4). For instance, isolates with tetracycline efflux pump gene tet(D) had higher doxycycline MICs, and those with sul and dfrA genes were predictably resistant to trimethoprim-sulfamethoxazole.
TABLE 3 Minimum Inhibitory Concentrations of the 5 KPC-Producing C. freundii Isolates

NOTE. Nonsusceptible or dose-dependent-susceptible minimum inhibitory concentrations per the Clinical and Laboratory Standards Institute (the US Food and Drug Administration for tigecycline) are bolded. AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CST, colistin; CTX, cefotaxime; DOX, doxycycline; ETP, ertapenem; FEP, cefepime; GEN, gentamicin; IMP, imipenem; KPC, Klebsiella pneumoniae carbapenemase; MEM, meropenem; SXT, trimethoprim-sulfamethoxazole; TGC, tigecycline; TOB, tobramycin; TZP, piperacillin-tazobactam.
TABLE 4 Antimicrobial Resistance Genes Identified by Whole Genome Sequencing

NOTE. The genes were identified by ResFinder at a cutoff of 80% similarity to the database entries. The genes had 100% identity unless specified. All strains possess bla KPC-3. bla CMY represents the chromosomal AmpC β-lactamase gene intrinsic to Citrobacter freundii.
By pulsed-field gel electrophoresis, isolates M1 and M2 showed an identical genomic restriction pattern, and isolate M3 was closely related to isolates M1 and M2 with a 3-band difference. On the other hand, isolates M4 and M5 had distinct restriction patterns (Figure 2). Multilocus sequence typing corroborated this result, with isolates M1-M3 belonging to the same sequence type (ST102), whereas isolates M4 and M5 each belonged to separate sequence types (Table 5). Whole genome phylogenetic analysis showed that isolates M1-M3 belonged to the same clone with a shared core genome (Figure 3, dots) whereas isolates M4 and M5 were unrelated to M1-M3 and to each other (Figure 3, squares). Furthermore, M1-M3 were very closely related with only 7-11 single-nucleotide polymorphism differences whereas M4 and M5 had more than 184,056-198,984 single-nucleotide polymorphism differences (Table 6).
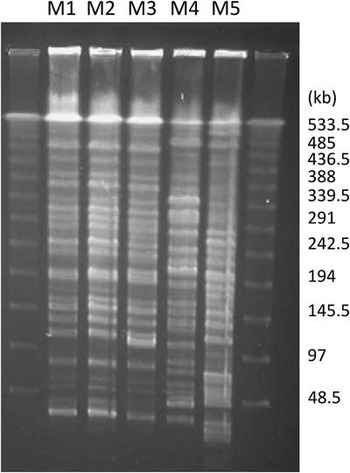
FIGURE 2 Pulsed-field gel electrophoresis of the 5 Klebsiella pneumoniae carbapenemase–producing Citrobacter freundii isolates.
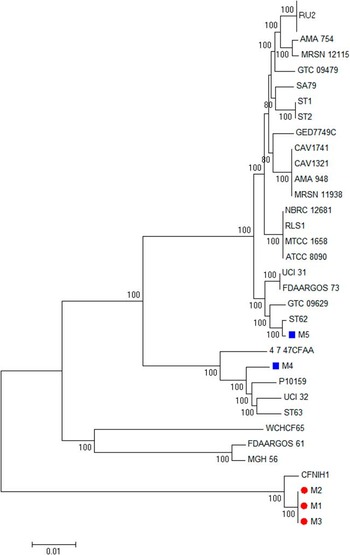
FIGURE 3 Core genome phylogenetic tree of Citrobacter freundii isolates. M1-M3 belonged to the same clone with a shared core genome (dots) whereas isolates M4 and M5 were unrelated to M1-M3 and to each other (squares).
TABLE 5 MLST alleles and STs of the 5 KPC-producing Citrobacter freundii isolates

NOTE. KPC, Klebsiella pneumoniae carbapenemase; MLST, multilocus sequence typing; ST, sequence type.
TABLE 6 Pairwise SNP Differences Among the 5 KPC-Producing Citrobacter freundii Isolates

NOTE. KPC, Klebsiella pneumoniae carbapenemase; SNP, single-nucleotide polymorphism.
These findings indicated that isolates M1 and M2 were identical or nearly identical, and isolate M3 was also genetically very close to isolates M1 and M2, supporting direct transmission of carbapenemase-producing C. freundii ST102 among these patients in SICU-B. Contrarily, isolates M4 and M5 were distinct from isolates M1-3 and from each other and represented sporadic cases despite temporal association within the same hospital.
DISCUSSION
Carbapenem resistance in the family Enterobacteriaceae is one of the greatest threats that are evolving in the field of medicine and public health at large, with the potential to roll back the gains of modern medicine owing to limited treatment options for infections. The most common type of CPE is KPC-producing Enterobacteriaceae, which initially caused outbreaks in hospitals in the mid-Atlantic United States in the early 2000s and has since spread across the United States and to other countries. KPC-producing Enterobacteriaceae is overrepresented by K. pneumoniae, which in turn accounts for the majority of CPE.Reference Alrowais, McElheny and Spychala 12
Prior to the outbreak, the facility in this report had a median of 7 patients with CPE present in the institution per day, but they were mostly K. pneumoniae, and C. freundii had not been identified earlier. We initially noticed an increased number of carbapenemase-producing C. freundii isolates in March 2015. This unusual finding alerted us to (1) enforce control measures in addition to the bundle already in place, and (2) prospectively monitor for additional cases. We included in this investigation all carbapenemase-producing C. freundii patients identified by reviewing the infection control databases from January 2014 to July 2015.
By carbapenem resistance and phenotypic carbapenemase testing, we were able to identify a total of 6 cases. Unfortunately the isolate of the first chronological case (M6) was not available for pulsed-field gel electrophoresis or further testing; this may have been a sporadic case, but it is also possible that it was related to M1-M3 and that it was actually the index case. The clinical and molecular investigation provided conclusive evidence that at least 3 of the 6 cases constituted transmission events because the patients were admitted in proximity in the same intensive care unit and the isolates were highly clonal by pulsed-field gel electrophoresis as well as multilocus sequence typing. On the other hand, at least 2 of the 3 remaining cases were sporadic on the basis of the investigation, and these isolates incidentally showed lower MICs of carbapenems by confirmatory testing despite carriage of the same carbapenemase gene bla KPC-3. A wide range of carbapenem MICs have been reported among non-K. pneumoniae, KPC-producing Enterobacteriaceae, including E. coli and C. freundii.Reference Deshpande, Rhomberg, Sader and Jones 16 A high degree of vigilance may therefore be required in detecting carbapenemase production in these species.
We were able to control the transmission of KPC-producing C. freundii thanks to the rapid detection of the outbreak, the bundle of interventions, and the enhanced environmental cleaning used to handle the outbreak among other measures taken. Unfortunately we did not collect environmental cultures that would support the benefit of enhanced cleaning; it is also possible that this outbreak would have waned without interventions, though this would not have been a practical option from an infection control standpoint. Moreover, the plumbing issues in the SICU may have negatively affected compliance with hand hygiene practices and facilitated patient-to-patient transmission of KPC-producing C. freundii. The extra environmental cleaning and disinfection likely played a pivotal role in arresting the spread of this CPE in our facility. Once the patients were moved to a terminally cleaned new pod, transmission of carbapenemase-producing C. freundii in SICU-B stopped. Interestingly, carbapenemase-producing C. freundii was not detected in the surveillance cultures of these patients. On the other hand, most of the carbapenemase-producing K. pneumoniae were first identified by surveillance cultures at our institution. This finding raises questions about the sensitivity of current surveillance methods to detect some carbapenem-resistant Enterobacteriaceae species other than K. pneumoniae, along with the fact that some of the isolates had relatively low carbapenem MICs. More sensitive methods for CPE surveillance culture may need to be explored as carbapenemase-producing non-K. pneumoniae species become more common.
We acknowledge several limitations to the investigation. The small number of cases limited generalizability of the findings, which reflected successful control of the outbreak. Also, we were unable to determine the source and mechanism of horizontal transmission in SICU-B where the defined outbreak cases occurred.
In conclusion, we report investigation, control, and molecular analysis of an outbreak of KPC-producing C. freundii at a tertiary care facility in Florida. Although C. freundii is not a common source of CPE, our findings suggest that carbapenemase-producing C. freundii may be underdetected even when active surveillance is in place and that it has the potential to cause a hospital outbreak with a negative impact on patient outcome and resource utilization.
ACKNOWLEDGMENTS
We thank Maribel Ruiz, for her assistance delineating the timeline of this outbreak; and Piotr Majewski, MD, for his assistance with curation of the new multilocus sequence typing alleles and sequence types.
Financial support. National Institutes of Health (grants R01AI104895 and R21AI123747 to Y.D.).
Potential conflicts of interest. L.S.M.-P. reports that she has served on an advisory board for Xenex and consulted for Clorex. Y.D. reports that he has served on advisory boards for Shionogi, Meiji, Tetraphase, and Achaogen, received a speaking fee from Merck, and received research funding from Merck and The Medicines Company for studies unrelated to this work. All other authors report no conflicts of interest relevant to this article.











