Acinetobacter baumannii is known to cause serious infections and has been associated with nosocomial exposure. Multidrug-resistant A. baumannii, specifically carbapenem-resistant A. baumannii (CRAB), has been identified as a serious threat by the Centers for Disease Control and Prevention (CDC).1 The vast majority of patients with CRAB infections have had recent exposure to the healthcare system, and mortality approaches 20%.Reference Bulens, Yi and Walters2 Known to prefer moist environments and to be resistant to desiccation, A. baumannii has been identified on hospital surfaces, in air samples, and less commonly in bulk water.Reference Wong, Nielsen, Bonomo, Pantapalangkoor, Luna and Spellberg3,Reference Shamsizadeh, Nikaeen, Nasr Esfahani, Mirhoseini, Hatamzadeh and Hassanzadeh4
Transmission of multidrug-resistant organisms such as CRAB in hospital settings can occur via multiple pathways by direct and indirect contact.Reference Blanco, O’Hara and Harris5 Pathogens have been cultured from terminally cleaned hospital rooms, and transfer between patient and environment has been demonstrated to occur early during admission.Reference Chen, Knelson and Gergen6
Between July 2018 and January 2019, we identified 5 cases of CRAB at the Massachusetts General Hospital (MGH). Four patients had been cared for sequentially in the same hospital room in the burn intensive care unit (BICU). A fifth patient was identified in a different ICU, the medical ICU (MICU). An extensive epidemiological investigation was conducted, including whole-genome sequencing (WGS), which linked all cases to the index case. We report on the findings of that investigation, including possible environmental reservoirs that may have contributed to transmission.
Methods
Identification of index case
During routine surveillance of microbiological results at our institution, the index case was identified in July 2018, on hospital admission day 25 (HD25). The index case represented the first CRAB isolated from a BICU patient in >3 years. Prior to admission, the index patient had no history of hospitalizations at our facility. The index patient was admitted only to the BICU and was on contact precautions beginning on HD2, when methicillin-resistant Staphylococcus aureus was isolated. The index patient had multiple clinical isolates of non–carbapenem-resistant A. baumannii complex beginning on HD7.
Identification of case patients
In a prospective review, 4 case patients were identified from clinical microbiology results. In October 2018, a BICU patient (case 1, C1) was identified with CRAB on HD2, while the index patient was admitted. In November 2018, a second BICU patient (case 2, C2) was identified with CRAB on HD2 while patient C1 was admitted. In December 2018, a third BICU patient (case 3, C3) was identified with CRAB on HD5 while patient C2 was admitted. Also, patient C3 was identified with CRAB 2 days after admission to the same room where the index patient, patient C1, and patient C2 had been previously admitted (Fig. 1). In January 2019, a fourth patient (case 4, C4) was identified with CRAB on HD24 in the MICU during routine review of clinical isolates.

Fig. 1. Investigation timeline. The Index (I) and Case patient (C1–C3) admission locations with date of CRAB first isolated indicated(*). C4 was identified in a different hospital unit and is not included here.
Investigation
The epidemiological investigation focused on the review of prior microbiology of the index patient and patients C1–C4, as well as identifiying any shared locations, procedures, and providers. Infection prevention practices on the BICU were assessed, and later in the MICU when patient C4 was identified. Direct observations of practices in the BICU, including daily and terminal cleaning of the room, as well as cleaning and disinfection of portable medical equipment (PME), were conducted. Environmental sampling was performed in the common patient room while patient C3 was admitted, and again after terminal cleaning. A subset of the environmental CRAB isolates underwent WGS.
Laboratory analysis of clinical samples
Clinical samples were tested at the MGH microbiology laboratory according to standard procedures. Bacterial isolates were identified using matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS; Vitek MS, v3.0, bioMérieux, Durham, NC). Routine antimicrobial susceptibility testing (AST) was performed using the Vitek 2 system (bioMérieux). Additional AST was performed as noted (Table 1).
Table 1. Characteristics and Antimicrobial Susceptibility Testing Profiles of Acinetobacter baumannii Isolates Obtained From Index Case and Case Patients

Note. CRAB, carbapenem-resistant A. baumannii; MIC, minimal inhibitory concentration; R, resistant; S, susceptible; I, intermediate; NI, no interpretation.
a For the index case isolate, amikacin and tobramycin susceptibility testing was performed using the Sensititre Gram-Negative GN4F MIC Plate and for the C1 isolate, andamikacin susceptibility testing was performed using the standard Clinical and Laboratory Standards Institute (CLSI) disk diffusion procedure (using BBL Sensi-Disc Susceptibility Test Discs and BBL Mueller-Hinton Agar; Becton Dickinson).
b The hospital day and specimen source are provided for the case isolate that underwent WGS. In most cases, A. baumannii complex was isolated from multiple body sites, but (when available) AST profiles across all patient isolates were essentially identical; thus, only 1 isolate per patient was analyzed using WGS.
c Quality-control organisms included Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Klebsiella pneumoniae ATCC 700603.
d Vitek 2 system.
e Sensititre broth microdilution method.
f Minocycline MICs were determined using either the Sensititre Gram-Negative GN4F MIC plate or the gradient diffusion method (Etest; bioMerieux) and ceftazidime-avibactam MICs were determined using Etest.
g Ellipses (…) indicate that testing was not performed.
h Gradient diffusion.
i Colistin and tigecycline MICs were determined using the Sensititre Gram-Negative Research Use Only GNX3F MIC Plates (TREK Diagnostic Systems; Cleveland, OH) that had undergone in-house performance validation.
j Disk diffusion.
Laboratory analysis of environmental samples
Environmental samples were tested at Microbiology Specialists (Houston, TX). A MacConkey broth (Hardy Diagnostics, Santa Maria, CA) was inoculated and incubated in ambient conditions at 35°C for 48 hours before being subcultured to MacConkey agar (Remel, Lenexa, KS) with ertapenem and meropenem disks (BBL Sensi-Disc, Becton Dickinson, Sparks, MD). Any resistant isolates were subcultured to a blood agar plate (Remel) then incubated at 35°C to ensure purity. Pure suspect isolates were identified by MALDI-TOF MS (MALDI Biotyper, Bruker Daltonics, Billerica, MA) using the Bruker database; species-level identification was confident for scores ≥2. Additional AST was performed using the standard Clinical and Laboratory Standards Institute (CLSI) disk-diffusion procedure using BBL Sensi-Disc Susceptibility Test Discs for meropenem, imipenem, and ertapenem; Hardy Susceptibility Test Discs for doripenem; and Mueller-Hinton agar (Remel). Quality-control organisms included Pseudomonas aeruginosa ATCC 27853. Non-CRAB organisms found within the screening zones of inhibition were also identified by MALDI-TOF MS.
Genome sequencing
Genomic sequencing and analysis were performed at Day Zero Diagnostics (Boston, MA). Genomic DNA from 10 samples (from the index case, C1–C4, and 5 environmental samples) was isolated from cultured isolates using organic aqueous-phase, bead-beating, crude extraction followed by a QiaAmp DNA Mini Kit (Qiagen, Venlo, Netherlands). Libraries were prepared with Nextera tagmentation (Illumina, San Diego, CA) and were amplified using unique, dual-indexed primers. An iSeq 100 instrument (Illumina) was used to generate 2×150-bp paired-end reads.
Genomic analyses
We performed SNP-based sequence analysis on the 10 isolates to measure their relatedness. We included in this analysis an additional 21 clinical A. baumannii isolates (10 of which were CRAB), collected from MGH between 2015 and 2018 to assess the relatedness of these isolates in the context of other MGH strains. De novo assemblies were generated using the SPAdes v3.13.0 Kbase application. Contigs <500 bp in length were removed. Multilocus sequence typing (MLST) was performed using assembled contigs by matching locus alleles to the PubMLST database. Sequencing reads were mapped to a circularized reference genome A. baumannii ATCC 17978 (GenBank: CP000521.1) using the BWA mem v0.7.13 algorithm. SNP calling was performed using the Pilon v1.23 tool. Only regions with read-mapping quality >30 and covered by >5 paired-end reads were considered. Insertions, deletions, mixed alleles (minor allele frequency ≥ 15%), and putative recombination regions (identified using the gubbins v2.3.1 package)Reference Croucher, Page and Connor7 were excluded from the final SNP analysis. Pairwise SNP distance was calculated using custom scripts. The phylogenetic tree was built using the RaxML v8.2.12 tool.Reference Stamatakis8
Because the sequence type of the reference genome differed from that of the isolates (no closed genome with a closely related MLST was available in NCBI), only 3.4 MB of the 3.9-MB reference genome was analyzed. Therefore, we performed a supplementary analysis by aligning the reads of each sample to the de novo assembly of the index case using the same SNP analysis pipeline as described above. This analysis achieved qualitatively similar results.
Human subjects
The activities conducted as part of this investigation were considered part of the routine infection control response at MGH, and submission to the Partners Human Subjects Committee was not required.
Results
Case identification and investigation
Cases were identified through routine surveillance for multidrug-resistant organisms. The A. baumannii isolates were reviewed by infection control staff due to their resistance to carbapenems but were also highly resistant to ampicillin-sulbactam, aminoglycosides, fluoroquinolones, tetracycline, and trimethoprim-sulfamethoxazole (Table 1). They were uniformly susceptible to colistin, had variable susceptibility to minocycline, and had tigecycline minimal inhibitory concentrations (MICs) <2 µg/mL. A review of CRAB dating back to 2013 revealed between 0 and 2 cases identified per month prior to this cluster (Fig. 2). The AST profiles of all CRAB isolates from January 2018 through May 2019 were reviewed and compared to the AST profiles of the index and case isolates. Isolates from 3 patients were indistinguishable from the index and case isolates on the basis of AST (P1, P2, and P3, for possible cases 1, 2, and 3); these isolates had been discarded and were unavailable for further analysis.

Fig. 2. Monthly, hospital-wide CRAB cases, January 2013 through January 2019. The index case (ᵻ) and cases C1–C4 (*) identified as part of this investigation are noted.
In addition to manual chart review, an electronic records extract inclusive of all patient locations, procedures, tests, and providers was performed, comparing the index patient and patients C1–C4. This query confirmed many overlapping interactions between the index patients, patient C1, patient C2, and patient C3, which was to be expected because all were admitted to the BICU. During this period, 4 additional patients were admitted to the same BICU room; no CRAB was identified in the review of clinical microbiology from those patients. A single commonality was identified through the electronic data extracted between patients C3 and C4 (the latter being identified in the MICU): both had exposure to the same PME. This extract identified multiple portable chest radiographs a single ultrasound in common; these procedures were conducted prior to the identification of CRAB in C4.
Infection control and environment assessment
Infection control evaluations included multiple on-unit observations to assess standard infection control practices such as hand hygiene compliance, implementation of transmission-based precautions, and cleaning of the patient rooms. An inventory of PME in use on the BICU was evaluated. There were no observed or known breaches in hand hygiene or use of personal protective equipment. The infection control and environment assessment focused on the room common to the index patient and patients C1–C3, as well as use of PME entering and exiting the room. An evaluation of the mattress revealed concern regarding the feasibility of adequate cleaning and disinfection of the sleeve used to insert portable film cartridges. Based on this assessment, the decision was made to require the use of disposable plastic sleeves for each portable film cartridge, in addition to the manufacturer’s recommendations regarding cleaning the sleeve. Several changes were made to bed-cleaning procedures, including review and re-education of staff and assignment of a minimum of 2 individuals to the terminal cleaning. The bed undergoes 2-step cleaning and disinfection to ensure both removal of bioburden and disinfection.
Laboratory analysis of environmental isolates
Multiple environmental samples obtained before cleaning were positive for CRAB (Table 2), including locations on the bed and mattress (including mattress sleeve), the overhead lift, the sink bowl and drain, and electrocardiography leads (Fig. 3). A subset of locations and items sampled in the precleaning period were resampled after C3 was discharged and the room was terminally cleaned. CRAB was not isolated from any of these samples, but other potentially pathogenic organisms were recovered from several locations.
Table 2. Environmental Sampling With Identification of CRAB and Other Organisms Before and After Cleaning
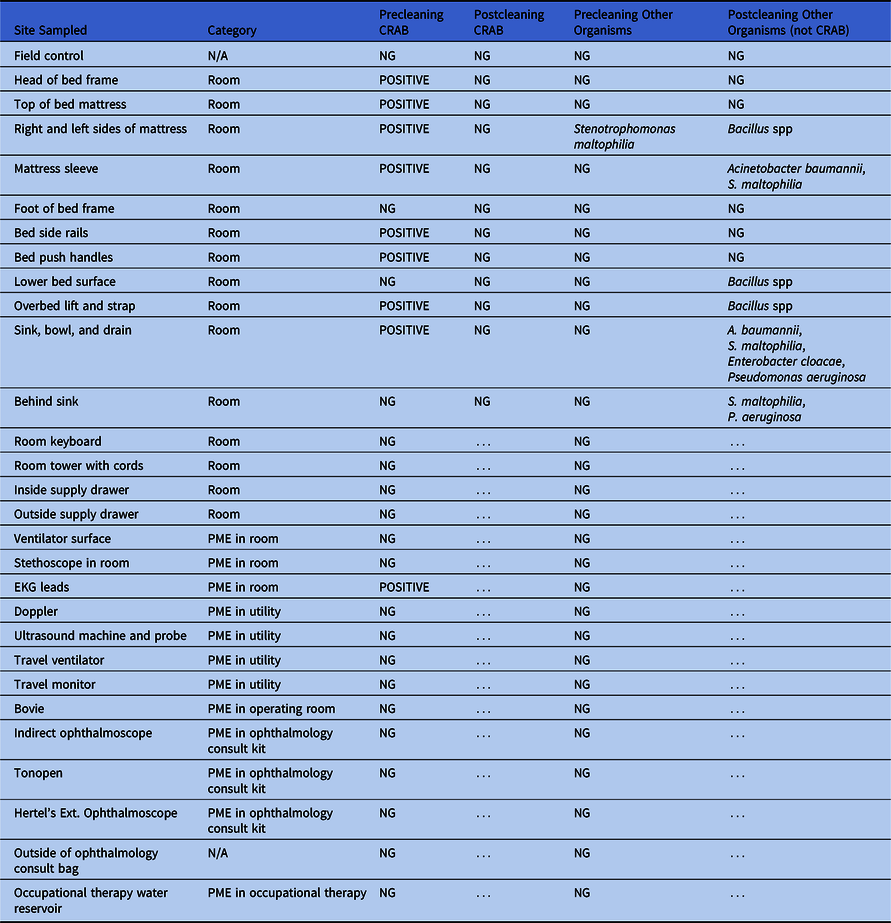
Note. CRAB, carbapenem-resistant A. baumannii; NG, no growth; PME, portable medical equipment; N/A, not available; EKG, electrocardiogram. … indicates testing not performed.

Fig. 3. Environmental sampling locations. Red circles represent locations sampled before cleaning that were contaminated with CRAB. Not all locations sampled are shown. Refer to Table 2 for details.
Genomic analysis of clinical and environmental isolates
Whole-genome sequencing was performed for 10 isolates (1 each from the index case and C1–C4, plus 5 environmental samples), yielding >190 MB and an average depth >50x per sample. The MLST analysis using the Oxford protocol revealed that all 10 samples were of sequence type ST105. The WGS reads from each isolate were aligned to an A. baumannii ATCC 17978 reference genome (GenBank no. CP000521.1), and SNPs were called for each sample.
Pairwise SNP differences between the A. baumannii isolates were called after putative recombination regions were excluded (Supplementary Table 1 online).Reference Fitzpatrick, Ozer and Hauser9 The small number of SNP differences indicates that all isolates were highly related. The 5 case isolates (index and C1–C4) were between 1 and 7 SNPs in pairwise SNP distance. Similarly, all case isolates were between 0 and 10 SNPs to the environmental isolates (Supplementary Table 1 online).
In contrast, WGS of an additional 21 A. baumannii clinical isolates, considered representative of background A. baumannii at the institution, demonstrated multiple divergent sequence types. Phylogenetic analysis revealed that the genetic diversity between A. baumannii background isolates was much higher than between the cluster isolates and that the background isolates were not closely related to the cluster isolates (Fig. 4). Most of the background isolates had a distance >20,000 SNPs from the cluster isolates. Two background isolates (background 1 and background 2) were of a MLST profile related to that of the cluster isolates, with only 1 allele difference from ST105. Nevertheless, the 2 background isolates had >130 SNP distance to the outbreak isolates.
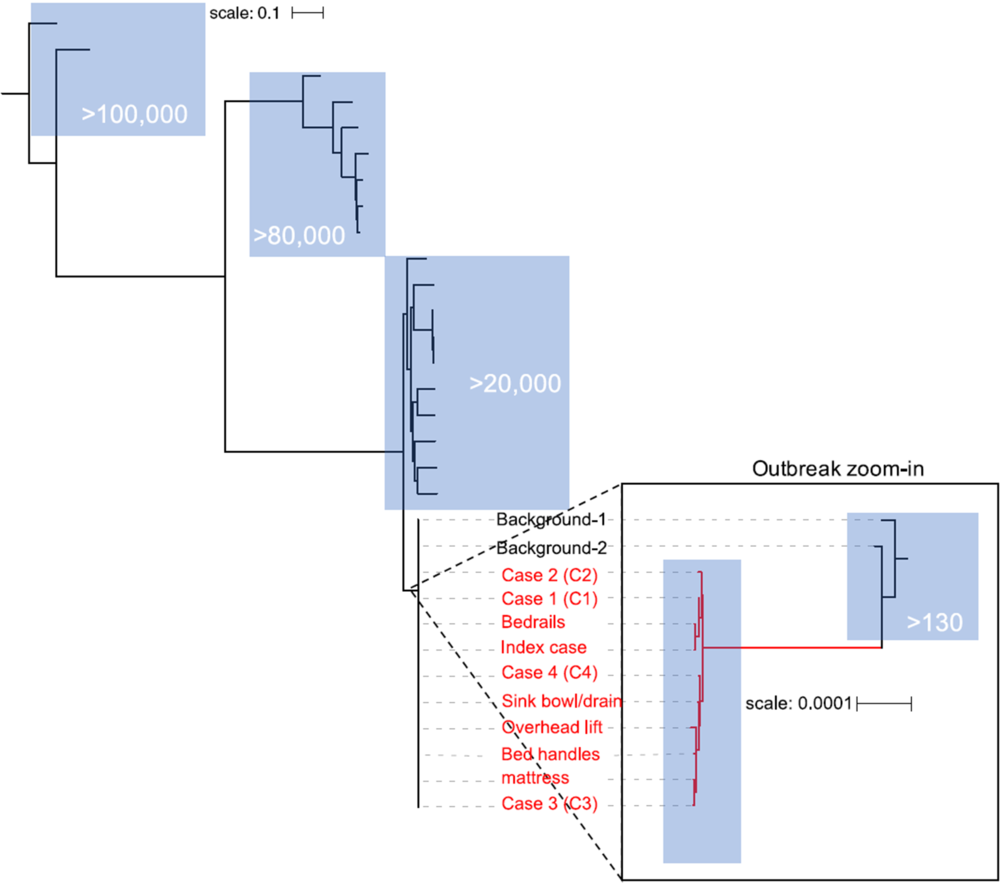
Fig. 4. Genetic diversity of cluster and background Acinetobacter baummanni isolates presented in a phylogenetic tree. The branch containing the outbreak isolates is colored in red. Blue boxes highlight clades with the corresponding rough SNP distance from the outbreak cluster clade (in white text).
Discussion
We describe an outbreak investigation of highly resistant A. baumannii. Although the investigation initially focused on patients in a single ICU, all of whom had been admitted to a common room in sequence, it eventually included a patient in a different ICU with no shared locations or providers. Environmental sampling revealed possible reservoirs.
Although the common room implicated in cases C1–C3 could have contained reservoirs that led to transmission, transmission via transient colonization of healthcare worker hands or personal protective equipment, and indirect contact across room locations within the BICU are possible as well, given that each patient overlapped temporally with the prior patient. Patients C1–C3 were likely highly susceptible to infection given their extensive burns and lack of skin integrity, which put them at high risk for rapid onset of infection in the setting of exposure. No obvious linkage between cases C3 and C4, in a different (though connected) building, 7 floors apart, could be established, with the exception of common exposure to PME. Other transmission pathways, including contamintation of common areas, such as elevators and handrails, could have resulted in transmission from patient C3 to C4. The BICU rooms present an additional challenge because they may be kept at elevated temperature and humidity levels to minimize patient heat and fluid loss, and these conditions improve the growth and survival of many bacteria, including Acinetobacter.Reference Jawad, Snelling, Heritage and Hawkey10,Reference De Silva, Chong, Fernando, Westmacott and Kumar11
This investigation has revealed insights into the challenges facing healthcare facilities in ensuring that the built environment and PME are adequately cleaned and disinfected to reduce the risk of transmission. Given the frequency with which healthcare workers interact with the patient, the patient’s environment, and PME,Reference Jinadatha, Villamaria and Coppin12,Reference Donskey13 as well as known failures in hand hygieneReference Clack, Scotoni, Wolfensberger and Sax14-Reference Longtin, Schneider and Tschopp16 and use of personal protective equipment,Reference Krein, Mayer and Harrod17-Reference Tomas, Cadnum and Mana19 there are many opportunities and potential pathways for transmission. The process for bed and mattress cleaning and disinfection is estimated to take an hour. This time includes only 1 piece of equipment in the room; a full room cleaning can take more time than may be currently allotted given the demand for bed availability. The use of PME that enters and exits a room is further challenged by the variety of device users and the need for clarity regarding responsibility for ensuring that the devices are appropriately cleaned and disinfected.Reference Suwantarat, Supple, Cadnum, Sankar and Donskey20
The results of this investigation highlight the limitations of standard epidemiological definitions used to attribute infections to community or hospital, as well as using attribution to specific units and time frames alone as clues to potential transmission events. Using the CDC National Health and Safety Network definition of healthcare-associated infection,21 only 2 of the 4 case patients would be classified as nosocomial, when in fact all 4 cases, through WGS linkage to the index case, proved nosocomial. The hospital stay of patient C4 overlapped with that of patient C3, yet they were in distinct locations and no evidence indicates that they were ever in the same ancillary location or shared common providers. Had the isolate not been sequenced, there would have been no epidemiological reason to believe that C4 was part of the outbreak. Notwithstanding the value of WGS in identifying links that may have otherwise remained obscure, the use of WGS in detecting outbreaks has limitations. Without banked isolates, historical putative transmission events cannot be retrieved when a cluster is recognized through prospective epidemiological surveillance activities. Furthermore, selecting a single isolate from each patient for WGS fails to capture potential for within-host diversity and can therefore inflate SNP distances or confound the identification of transmission patterns. In this investigation, the small number of SNP differences between patient isolates and between patient and environmental isolates indicates that they were highly related, which was interpreted as evidence of transmission. Indeed, a recent study found A. baumannii isolates collected from the same individual to differ by as many as 11 SNPs.Reference Ng, Marimuthu and Lee22 However, the analysis presented here does not distinguish whether the outbreak was the result of expansion from a single strain or, less likely, the result of the introduction of multiple highly related strains (eg, from an environmental reservoir).
Such investigations are often triggered by unusual microbiological resistance patterns. At our institution, ~85% of A. baumannii complex isolates that undergo AST are susceptible to meropenem, and carbapenem-resistant A. baumannii complex are among the multidrug-resistant organisms for which infection control surveillance is routine. The isolates comprising this outbreak were also resistant to multiple other antimicrobials to which resistance at our institution is similarly uncommon (eg, ampicillin-sulbactam, gentamicin, and trimethoprim-sulfamethoxazole), and this pattern informed our decision to pursue WGS of the culture from C4 despite the lack of epidemiological links to the BICU cases. Institutionally uncommon AST profiles are a frequent trigger of infection control investigations and, in some instances, even a single rare organism could prompt investigation. However, given the abundance of putative transmission pathways, a substantial number of transmission events, especially those with more common organisms and resistance patterns, likely go undetected. Most genomically linked clusters are not detected using routine surveillance methods.Reference Ward, Hoss and Kolde23
The use of WGS for large-scale prospective surveillance to track pathogen transmission events has shown potential to alter the way outbreaks are identified and controlled. Real-time sequencing of recent viral oubreaks of EbolaReference Quick, Loman and Duraffour24 and ZikaReference Quick, Grubaugh and Pullan25 has demonstrated the utility of sequencing for public health epidemiological surveillance. The global laboratory PulseNetReference Nadon, Van Walle and Gerner-Smidt26 and the CDCReference Jackson, Tarr and Strain27 now advocate for real-time WGS of foodborne pathogens for disease surveillance. In clinical settings, although a number of pilot studies have implemented prospective WGS on large-scale collections of bacterial clinical isolates to identify transmission events,Reference Peacock, Parkhill and Brown28,Reference Mellmann, Bletz and Böking29 this approach has yet to be widely adopted. A more thorough understanding of the patient’s health improvements and the cost–benefits of using WGS to detect outbreaks like the one described here could help guide protocols that set the scope and implementation of WGS surveillance in hospital settings, including the most efficient approach to routine banking of clinical isolates. One challenge will be understanding the role of patient colonization, as well as environmental reservoirs (including the built environment and PME) in pathogen transmission; another is designing approaches that incorporate these reservoirs into analyses. Targeted sampling, made possible through approaches leveraging electronic health data, could help to focus sequencing efforts.Reference Ward, Hoss and Kolde23,Reference Sundermann, Miller and Marsh30
Acknowledgments
The authors wish to thank John Schulz, MD, PhD, Jonathan Friedstat, MD, MPH, Jeremy Goverman, MD, Robert Sheridan, MD, Jennifer Albert, RN, MS, CCRN, Anthony DiGiovine, RN, BSN, NE-BC, from the MGH Burn Intensive Care Unit; Suzanne Algeri, MS RN NE-BC, Associate Chief Nurse, MGH; George Reardon, MBA, Scott Parsons, MBA, and Awilda Lalande, MBA, Patient Care Services, MGH, for assistance in evaluation of cleaning and disinfection procedures; Marlene L. Durand, MD, Director of Hospital Epidemiology, and Debbie Rich, RN, CIC, Infection Preventionist, Massachusetts Eye and Ear Infirmary, Boston, Massachusetts, for assistance in evaluation of ophthalmologic PME; Alice S. Weissfeld, PhD, D(ABMM), F(AAM), Paula H. Vance, RM/SM(NRCM), SM(ASCP), CLS(M), CIE, Ernest Trevino, MT(ASCP), Fran Schaeffer, SM(ASCP), Rhonda Halliday, MT(ASCP), Hannah Livesay, M(ASCP), and Brooks Kennedy, BS, BCLS, MLS(ASCP) of Microbiology Specialists Inc, Houston, Texas, for assistance with analysis of environmental samples. The findings and conclusions in this report are those of the authors. Day Zero Diagnostics received a fee-for-service payment from Massachusetts General Hospital for the genomic sequencing and basic SNP analysis of the primary isolates. Additional research and analyses, content writing, and manuscript review were completed without payment.
Financial support
This work received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflicts of interest
M.R.A.S., F.K.P., I.C.H., and M.H.H. are employees of Day Zero Diagnostics. M.N.A. and D.S.K. are consultants to and hold equity in Day Zero Diagnostics. Massachusetts General Hospital holds a minor equity interest in Day Zero Diagnostics.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2020.15








