Diagnosis of Clostridium difficile infection (CDI) poses a unique challenge to clinicians because detection of C. difficile from a stool specimen alone does not distinguish between colonization and CDI.Reference Burnham and Carroll 1 The “gold standard” method for C. difficile toxin detection in stool specimens, the cell culture cytotoxicity neutralization assay, is slow; toxin enzyme immunoassays (EIAs) are faster but less sensitive. Nucleic acid amplification tests (NAATs) are both sensitive and fast; however, because NAATs have greater analytical sensitivity to detect C. difficile, NAATs may have poor positive predictive value (PPV) for CDI.Reference Polage, Gyorke and Kennedy 2
There is growing consensus that CDI diagnosis must combine laboratory results with appropriate clinical criteria, including presence of clinically significant diarrhea (CSD) and absence of other causes of diarrhea or worsening of diarrhea beyond what might otherwise be expected.Reference Dubberke and Burnham 3 – Reference Casari, De Luca, Calabro, Scuderi, Daleno and Ferrario 6 To this end, the recently released 2017 Infectious Diseases Society of America (IDSA)/Society for Healthcare Epidemiology of America (SHEA) Clinical Practice Guidelines for CDI update emphasizes that the “preferred” patient population for C. difficile testing is patients with unexplained new-onset CSD, defined as ≥3 unformed bowel movements within 24 hours. If testing cannot be limited to the preferred testing population, the use of a stool toxin test as part of a multistep diagnostic algorithm is the preferred method of testing, and NAAT should not be used alone.Reference McDonald, Gerding and Johnson 7 , Reference McDonald, Gerding and Johnson 8 If it is possible to limit testing to the preferred testing population, it is recommended a NAAT alone can be an acceptable testing method, but an acceptable alternative is the use of a stool toxin test as part of a multistep algorithm. Data are limited, however, on the prevalence of C. difficile colonization among the preferred testing population; this prevalence may affect the PPV of NAATs for CDI. Thus, the first step toward determining the PPV of NAATs for CDI is to address the knowledge gap surrounding the prevalence of C. difficile colonization among patients who meet the IDSA/SHEA guideline–preferred patient population for C. difficile testing.
To address this need, a retrospective cohort was assembled of patients with stool specimens tested with a toxin EIA during routine clinical care from patients with documented CSD and no identifiable alternate causes of diarrhea. These stool specimens were cultured for toxigenic C. difficile, and C. difficile isolates underwent polymerase chain reaction (PCR) ribotyping to determine the prevalence of C. difficile colonization among patients who met the IDSA/SHEA CDI clinical guidelines for the preferred C. difficile testing population.
Methods
An aliquot from each stool specimen submitted to the Barnes-Jewish Hospital (BJH, St. Louis, MO) clinical microbiology laboratory for C. difficile testing has been collected and stored at −80°C, quantity pending, since August 2014 for quality improvement purposes. Patients with stool specimens submitted for C. difficile testing at BJH from August 2014 through September 2016 by toxin EIA (Alere TOX A/B II, Abbott, Lake Bluff, IL) were eligible for inclusion. If a patient had >1 eligible stool specimens during an admission, the first stool specimen collected was used. Per hospital policy, only stool specimens that conformed to the shape of the container were tested for C. difficile. In May 2015, a restriction was placed on repeat testing if the patient had had a negative toxin EIA in the previous 4 days. The Washington University Human Research Protection Office approved this study.
Patients who did not have CSD or who had a potential alternate cause of diarrhea at the time the stool specimen was submitted for C. difficile testing were excluded. The hospital’s medical informatics database was queried to obtain toxin EIA results, microbiology results for detection of non–C. difficile enteric pathogens from stool specimens, International Classification of Disease, Ninth Revision, Clinical Modification (ICD-9-CM) and International Classification of Disease, Tenth Revision, Clinical Modification (ICD-10-CM) procedure and diagnosis codes, and medications administered. Electronic data were used to exclude patients if they had the following potential alternate causes of diarrhea: history of inflammatory bowel disease, irritable bowel syndrome, or colectomy in the previous 10 years; chemotherapy in the 14 days before stool specimen collection; hematopoietic cell transplant in the 180 days before stool specimen collection; alternate gastrointestinal pathogen isolated from the stool specimen up to 7 days before or after the stool specimen selected for this project was collected; tube feeds in the 48 hours before collection or presence of an ostomy, ileostomy, or ileal conduit; laxatives within 24 hours of specimen collection; previous history of CDI; or CDI antimicrobial treatment in the 10 days before and after specimen collection (eg, oral vancomycin, oral or IV metronidazole, or oral fidaxomicin) for EIA negative (EIA−) stools only. All patients with EIA+stool specimens received CDI treatment. Patients with an EIA− stool specimens who received CDI antimicrobial treatment within 10 days were excluded to ensure that patients with EIA−/toxigenic culture+stool specimens did not have CDI, which is the primary concern in using NAATs to aid in the diagnosis of CDI. Manual chart review was performed for all patients whose stool specimens were not excluded during electronic screening to determine whether the patients had any exclusion criteria that did not appear in the electronic data and whether the patients had CSD. All patients with CSD and no alternate cause of diarrhea were included in the study. Patients with EIA+/toxigenic culture+stool specimens were considered to have CDI, and patients with EIA−/toxigenic culture+stool specimens were considered colonized and without CDI.Reference Polage, Gyorke and Kennedy 2 , Reference Dubberke, Han and Bobo 9 – Reference Planche, Davies and Coen 12
Stool specimens were cultured for C. difficile according to previously published methods.Reference Hink, Burnham and Dubberke 13 Briefly, a 1-g aliquot of the stool specimen was heat shocked at 80°C for 10 minutes, inoculated into cycloserine-cefoxitin mannitol broth with taurocholate and lysozyme (Anaerobe Systems, Morgan Hill, CA), and incubated anaerobically at 35°C. When turbid, the broth was inoculated onto pre-reduced blood agar (BAP, Becton Dickinson, Franklin Lakes, NJ). Clostridium difficile colonies were identified using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) with the Vitek MS platform (bioMerieux, Durham, NC). Clostridium difficile isolates were evaluated for the presence of tcdA, tcdB, and binary toxin genes (cdtA/cdtB) by multiplex PCRReference Alasmari, Seiler, Hink, Burnham and Dubberke 14 – Reference Westblade, Chamberland and MacCannell 16 and underwent ribotyping. The ribotyping banding patterns were analyzed using DiversiLab Bacterial Barcodes software (bioMerieux). Strains were compared with the Cardiff-ECDC collection of C. difficile strains for name assignment. Strains without a match in the Cardiff-ECDC collection were assigned a Washington University (WU) strain number.
Results
A total of 8,931 patients had stool specimens tested for C. difficile during the study period (Fig. 1). Of these, 570 (6%) were EIA positive (EIA+) and 8,361 (94%) were EIA negative (EIA−). The electronic screening process excluded 293 EIA+stool specimens and 5,809 EIA− stool specimens with potential alternate causes of diarrhea, leaving 277 EIA+and 2,552 EIA− stool specimens eligible for chart review. The chart review process found an additional 169 EIA+stools and 2,037 EIA− stool specimens from patients whose alternate cause of diarrhea was identified or for whom study investigators were unable to find documentation of CSD.
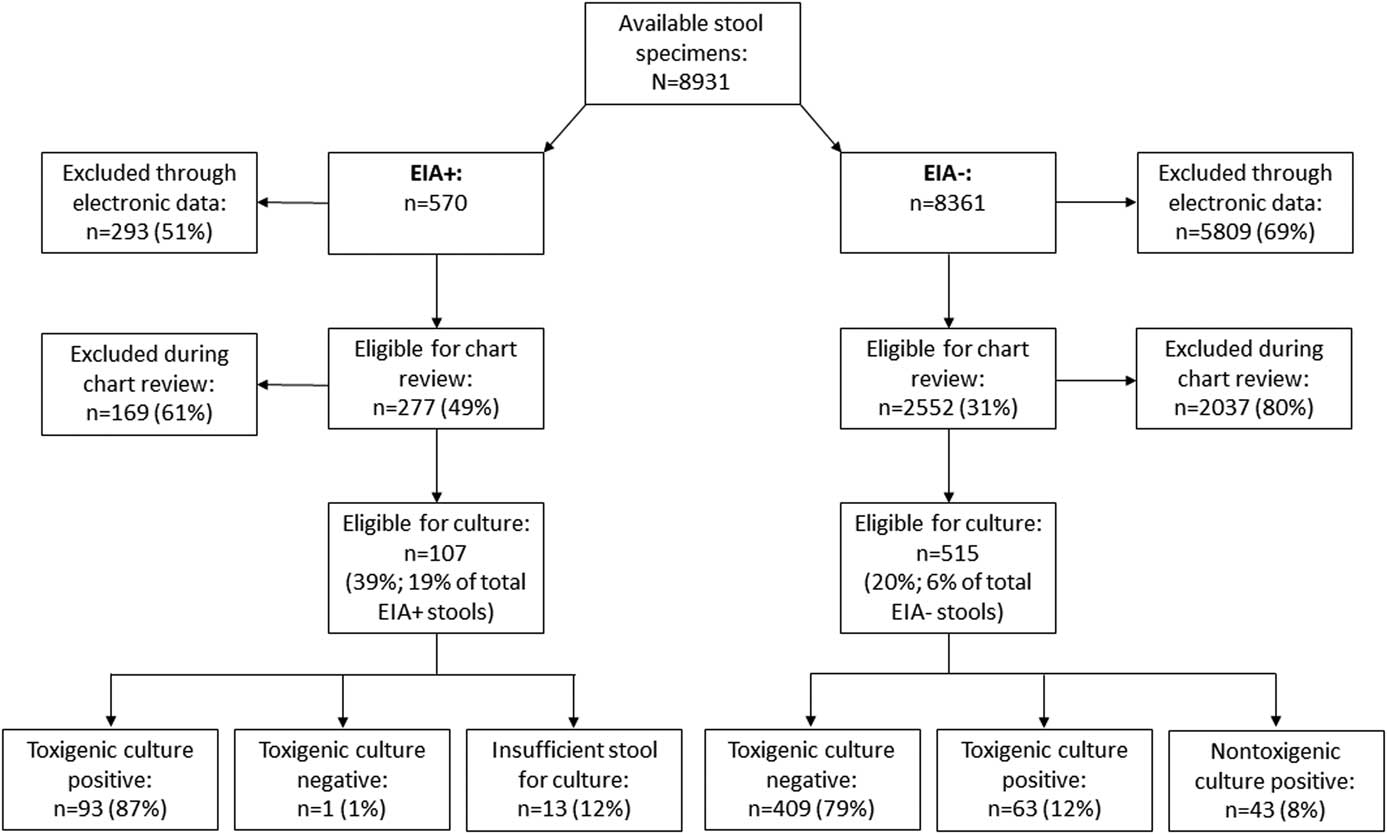
Fig. 1 Results of screening and C. difficile toxigenic culture.
The remaining 107 EIA+stool specimens (19% of all EIA+stool specimens) and 515 EIA− stool specimens (6% of all EIA− stool specimens) met inclusion and exclusion criteria and were cultured for C. difficile. Among the EIA+stool specimens, toxigenic C. difficile was isolated from 93 stool specimens (87%); 13 stool specimens (12%) could not be cultured due to insufficient stool volume; and 1 stool specimen (1%) was toxigenic culture negative. Among the EIA− stool specimens, C. difficile was not isolated from 409 stool specimens (79%), nontoxigenic C. difficile was isolated from 43 stool specimens (8%), and toxigenic C. difficile was isolated from 63 stool specimens (12%). Binary toxin was identified in 41 EIA+/toxigenic culture+isolates (44%) and 9 EIA−/toxigenic culture+isolates (14%). The 3 most common strains isolated from EIA+stool specimens were ribotype 027 (n=35, 38%), ribotype 106/174 (n=14, 15%), and ribotype 002 (n=10, 11%). The 3 most common toxigenic strains isolated from EIA− stool specimens were ribotype 014/020 (n=10, 16%), followed by ribotype 027 (n=8, 13%); ribotype 001 (n=6, 10%), and ribotype 106/174 (n=6, 10%). None of the patients with EIA−/toxigenic culture+stool specimens had a documented microbiologically or clinically confirmed diagnosis of CDI within 30 days of the negative EIA.
Discussion
Using very strict criteria to ensure no potential alternate causes of diarrhea, only 622 stool specimens (7%) submitted for C. difficile testing were from patients with documented CSD and met the IDSA/SHEA criteria for the preferred C. difficile testing population.Reference McDonald, Gerding and Johnson 7 , Reference McDonald, Gerding and Johnson 8 Among the stool specimens that were EIA+, 19% met these criteria; among EIA− stool specimens, only 6% did. Not surprisingly, toxigenic C. difficile was isolated from 93 of 94 (99%) EIA+stool specimens with sufficient stool for culture. This finding suggests that toxin EIA has excellent specificity for detecting patients with CDI among patients who meet the preferred C. difficile testing population definition. The most common strain isolated from EIA+stool specimens was ribotype 027, which was identified at a proportion similar to that seen in 2010 at BJH.Reference Alasmari, Seiler, Hink, Burnham and Dubberke 14 The 027 strain accounted for 35 of 41 strains (85%) isolated from EIA+stool specimens with binary toxin. Toxigenic C. difficile was isolated from 63 (12%) of patients with EIA− stool specimens. Compared with EIA+/toxigenic culture+stool specimens, C. difficile isolated from EIA−/toxigenic culture+stool specimens were less likely to have binary toxin. The strain distribution of toxigenic isolates from EIA− stool specimens was different than that from EIA+stool specimens. Ribotype 014/020 was the most common strain (16%), and ribotype 027 was the second most common strain (13%).
Most importantly, although all patients included in this study met the IDSA/SHEA criteria for the preferred C. difficile testing population, the recovery of toxigenic C. difficile from these EIA− stool specimens likely represented colonization and not CDI. Patients were excluded if they received empiric treatment for CDI (or were on metronidazole for other reasons), and no EIA−/toxigenic culture+patients were diagnosed with CDI within 30 days of when the stool specimen selected for this study was collected. The sensitivity of NAAT for detecting C. difficile from diarrheal stool specimens submitted for C. difficile testing compared to toxigenic culture in general has been found to be ~90% to 100%.Reference Crobach, Baktash, Duszenko and Kuijper 17 Toxigenic C. difficile was isolated from 156 stool specimens, 63 of which were EIA−. Excluding the EIA+stool specimens without sufficient stool for culture and assuming that all NAATs would be positive from all EIA+/toxigenic culture+stool specimens, presumptively the PPV of NAAT for CDI from this population would be 60%–64%, similar to the PPV of NAATs for CDI seen when clinical presentation is taken into account.Reference McDonald, Gerding and Johnson 7 , Reference McDonald, Gerding and Johnson 8 , Reference Crobach, Baktash, Duszenko and Kuijper 17 Even if it were assumed that toxigenic C. difficile would have been recovered from the 13 EIA+stool specimens with insufficient stool for culture, the presumptive PPV of NAAT for CDI would be 63%–66%.
This study had several limitations. All stool specimens were initially submitted for C. difficile testing by clinicians and thus may have been subject to selection bias. As a retrospective study, it is possible that alternate, identifiable explanations for diarrhea could have been missed. Based on the number of patients excluded, we think that is unlikely. Notably, our definition for an alternate explanation for diarrhea was very broad because we felt it was more important to exclude a patient who did not necessarily have an alternate explanation for diarrhea than risk misclassifying a patient in the other direction. Although our exclusion criteria were very restrictive, they highlight the challenge of restricting C. difficile testing to patients who meet the preferred testing population. We were also unable to determine how many patients were excluded based on the lack of CSD alone; many patients had >1 reason for exclusion or were excluded using electronic data, from which CSD could not be determined. Another limitation of the definition is that patients can simultaneously have CDI and an alternate explanation for diarrhea. Also, we did not conduct NAATs, so the PPV of an NAAT among patients who met the IDSA/SHEA guideline–preferred C. difficile testing population criteria cannot be established and is based on conjecture. However, the sensitivity of the NAAT for detection of C. difficile from diarrhea stool specimens submitted for C. difficile testing compared to toxigenic culture is well established, and this study was not large enough to definitively establish the PPV of NAAT among patients who meet the IDSA/SHEA guideline–preferred C. difficile testing population criteria.
In conclusion, the IDSA/SHEA CDI clinical guideline update recommendations for C. difficile testing are stratified based on whether testing can be restricted to patients with unexplained and new-onset CSD. The NAAT is recommended as a stand-alone test if testing can be restricted to this population. The purpose of this stratification is because NAATs can detect C. difficile colonization among patients with diarrhea for other reasons. However, we found that 12% of patients who met strict criteria for new-onset CSD and no identifiable alternate cause of diarrhea were colonized with C. difficile. Additional research is needed to determine the optimal role for NAAT testing when there is clinical concern for CDI and/or if other markers can differentiate between C. difficile colonization and CDI.
Acknowledgments
Financial support
This work was supported by the Centers for Disease Control and Prevention’s investments to combat antibiotic resistance under award number 200-2016-91939. Dr Kwon reports that the research reported in this publication was supported by the Washington University Institute of Clinical and Translational Sciences (grant no. UL1TR000448) and a subaward (grant no. KL2TR000450) from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH.
Conflicts of interest
Outside the submitted work, E.R.D. reports grants from Rebiotix; grants and personal fees from Pfizer and Merck; and personal fees from Valneva, Rebiotix, Achaogen, Biofire, Abbott, and Synthetic Biologics. Outside the submitted work, C.D.B. reports grants from bioMerieux, Cepheid, and Luminex; grants and personal fees from Accelerate Diagnostics; and personal fees from BioRad and the Journal of Clinical Microbiology. All other authors report no conflicts of interest.



