Introduction
The analysis of the sex-disaggregated data of the ongoing pandemic of coronavirus disease 2019 (COVID-19) at a global scale shows that development of severe disease symptoms along with the mortality count is higher in men. A recent study has demonstrated clear difference in sex-based immunological response in COVID-19 (Ref. Reference Takahashi1). Scientists are still unravelling the unique properties of novel coronavirus virus strain causing this pandemic – severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). However, biological sex bias in the patient morbidity and mortality is not unique for the SARS-CoV-2, and similar antecedents have been known for previous virus infections, including from coronaviruses, such as severe acute respiratory syndrome (SARS) and middle-east respiratory syndrome (MERS) (Ref. Reference Channappanavar2). COVID-19-specific research explaining molecular mechanism(s) for sex-based differences in immunological responses is yet to be carried out. Key explanation for sex-biased presentations in COVID-19 is received from X-linkage of angiotensin-converting enzyme-2 (ACE2) gene, the human cell entry receptor for SARS-CoV-2 – which binds to viral spike protein. ACE2 is an interferon (IFN)-stimulated gene (ISG), which in turn has an oestrogenic regulation at the gene level (Refs Reference Scully3, Reference Bunders and Altfeld4). Similarly, a human protease, transmembrane protease, serine 2 (TMPRSS2), which primes viral spike protein for host cell entry, has an androgenic regulation at the gene level (Refs Reference Scully3, Reference Bunders and Altfeld4). Several immunoresponsive genes, such as Toll-like receptors 7 and 8 (TLRs-7 and -8), interleukins viz. IL-4, IL-10, IL-13, FoxP3 and CD-40L are X-linked, which are crucial in priming immune response against viral infections including coronaviruses (Refs Reference Scully3, Reference Bunders and Altfeld4). However, literature cites multiple additional mechanisms in relation to other infectious diseases which may hold true for COVID-19, such as evolutionary existence of stronger immune response in the case of females (Refs Reference Úbeda and Jansen5, Reference Zuk, Stoehr, Klein and Roberts6), immunomodulatory effect of sex hormones (Ref. Reference Taneja7) and gut microbiome (Ref. Reference Dhar and Mohanty8). In this paper, we have reviewed the existing empirical knowledge about the molecular mechanisms behind sex bias in immune responses against the respiratory viruses, with a focus on COVID-19.
Materials and methods
Objective
To identify plausible molecular mechanisms implicated in sex-based differences in patient outcomes in COVID-19.
Information sources and search strategy
The online literature sources including PubMed, Medline (EBSCO and Ovid), Google Scholar, ScienceDirect, Scopus, BioMedical and Web of Science (WoS) were explored extensively for the relevant data. Additionally, publication citations were also searched. The time period taken to review the COVID-19-specific data included from 1 December 2019 to 15 December 2020. The terms used for the search included COVID-19 and sex/sex differences, sex-based morbidity/mortality in COVID-19, molecular mechanisms, evolutionary basis, testis/ovary pathology in COVID-19, ACE2 expression in reproductive organs, immunological basis, genetic basis, epigenetics, role of smoking, microbiome, etc., plus months and year of study.
Protocol followed
A systematic review of recent studies was performed following Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Supplementary file 1).
Data selection, extraction and qualitative synthesis
Full articles which provided information about sex-based morbidity/mortality, mechanism of entry of the SARS-CoV-2 virus and histological changes in reproductive tissue, sex hormone alterations, immunological responses in COVID-19 patients were included in the study. The original clinical/epidemiological/animal model studies showing significant statistical associations [in terms of odds ratios (ORs) and/or risk ratios] of biological sex of the patients/in vivo models with morbidity and mortality rates in COVID-19 or other respiratory viruses (SARS, MERS and influenza) were considered for qualitative analysis. Additionally, relevant non-COVID articles explaining evolutionary, genetic and epigenetics underlying sex-based differences in immunological response against respiratory viruses were also included. Full length original articles, case studies and meta-analysis papers were included. The databases presenting expression of SARS-CoV-2 host cell entry receptors and proteins in human tissues, and COVID-19 demographic details, were reviewed for the sex-differentiated data. In addition, articles presenting commentary and review of the current COVID-19 research were scrutinised for relevant descriptions and original data links. A pre-publication peer review was conducted for the original articles in preprint (without peer review) to consider any of them for inclusion. The articles with only abstract and newsletters were excluded from the study.
For the original studies included in this review, any case was considered having COVID-19 only if tested positive for SARS-CoV-2 m-RNA in reverse transcription-polymerase chain reaction irrespective of the severity of the clinical symptoms. Complete data collection and qualitative analysis was contributed and reviewed by all the members of the research team, and disagreements among the investigators were resolved by recheck for the errors (in the data collection and analysis process) and mutual discussion. Any study with odd results has been discussed separately. The final inferences were made purely based on the qualitative synthesis from the collected data and no further quantitative data analysis or statistical testing was performed.
Results and discussion
Overall, 158 articles and databases were identified that fitted to our selection criteria. After exclusion of duplicates, 140 research articles were finalised for screening, following which 39 articles were excluded because of various reasons (duplication of information, no relevant data related to sex differences, etc.) and only 91 articles were found eligible for the final analysis (Supplementary file 1). Additionally, two databases [Human Protein Atlas (https://www.proteinatlas.org/humanproteome/sars-cov-2) and Global Health 50/50 (https://globalhealth5050.org)] were used, which presented relevant sex-differentiated data. Articles were grossly categorised into following groups: providing details for (i) sex-specific prevalence, (ii) cell entry receptor-based mechanisms, (iii) evolutionary and genetic bases, (iv) role of epigenetics, (v) hormonal basis, (vi) immunological basis and (vii) and additive factors, such as smoking and gut microbiome. We discuss below, salient observations from the COVID-19-specific studies under each category, in light of similar evidence received for the other respiratory viruses, wherever applicable.
Sex-specific prevalence of morbidity/mortality in COVID-19
Most of the studies, including meta-analyses, have shown higher prevalence of severe COVID-19 and mortality in men in comparison with women. Jin et al. studied sex-based differences in disease severity and mortality in patients with COVID-19 in (1) a case series of 43 hospitalised patients and (2) a public data set of the first 37 cases of patients who died of COVID-19 and 1019 patients who survived. Investigators also studied the archived data of 524 patients with SARS, including 139 deaths, from Beijing in early 2003. These investigators observed that older age and a high number of comorbidities were associated with higher disease severity and mortality in men as well as in women patients with COVID-19 and SARS. In the case series of patients under treatment, men's cases tended to be more severe compared with women's (P = 0.035). In the public data set, men and women had the same prevalence for contracting infection; however, the number of men who died from COVID-19 was 2.4 times that of women (70.3 versus 29.7%, P = 0.016). Sex bias in mortality was also observed in SARS patients, wherein, the percentage of men were higher in the deceased group than that in the survived group (P = 0.015) (Ref. Reference Jin9).
Williamson et al. conducted a cohort study using national primary care electronic health record data on 10 926 COVID-19-related deaths in the UK. Investigators found that, 90 days after the start of the study, the overall cumulative incidence of COVID-19-related death was less than 0.01% in those aged 18–39 years; however, in those aged 80 years or over, it was 0.67 and 0.44% in men and women, respectively. Men had a higher risk of death compared with women (fully adjusted hazard ratio 1.59 (1.53–1.65) (Ref. Reference Williamson10).
The meta-analysis of sex-differentiated data at a global scale robustly confirms that men are more vulnerable for developing severe disease and share higher mortality in comparison with women when caught with COVID-19. Recently, Peckham et al. analysed clinical data of 3 111 714 cases of COVID-19 from the published reports across the globe. The investigators observed that although there was no difference in the proportion of men and women with confirmed COVID-19, men had almost three times the odds of requiring intensive care unit (ICU) admission [OR = 2.84; 95% confidence interval (CI) = 2.06, 3.92] and higher odds of death (OR = 1.39; 95% CI = 1.31, 1.47) compared with women (Ref. Reference Peckham11). Another meta-analysis study by Yanez et al. analysed 178 568 COVID-19 deaths from a total population of approximately 2.4 billion people across 16 countries. The investigators observed that mortality rates from COVID-19 were 77% higher in men compared with those in women [incident rate ratio (IRR = 1.77; 95% CI = 1.74, 1.79] (Ref. Reference Yanez12). A further meta-analysis study by Nasiri et al. evaluating 5057 cases showed that the pooled mortality rate was on average 3.4 times higher in men as compared with women (95% CI = 1.2, 9.1; P = 0.01) (Ref. Reference Nasiri13).
The analysis of sex-disaggregated epidemiological data at a global scale involving 83 995 185 confirmed cases of COVID-19 from 145 countries does support a male sex bias in fatality reflected across the studies which we have discussed above (Fig. 1) (on 21 April 2021, source: https://globalhealth5050.org) (Ref. 14). Of note, against the global trend, a report cited a higher rate of severity of symptoms in women in India and Vietnam (Ref. Reference Kumar15) and also official reporting of sex-differentiated data from both of the countries reflect this (Ref. 14). However, for the countries, such as Nepal and Slovenia, which were earlier showing this trend are now catching up to the global trend with increased reporting of the sex-differentiated data (Ref. 14). Possibly, rather than a biological factor, an early reporting based on smaller sample size, limited official reporting of the sex-differentiated data and country-specific socio-demographic factors might have been responsible for the aberrant reporting (Ref. Reference Dehingia and Raj16).

Fig. 1. COVID-19 sex-disaggregated data at a global scale. Data source: The COVID-19 Sex-Disaggregated Data Tracker, Global health 50/50 (https://globalhealth5050.org, an open access database. The data were reviewed until 21 April 2021).
Cell entry receptor-based molecular mechanism(s)
SARS-CoV-2 entry in human cells is mediated by a cell surface protein ACE2 (Ref. Reference Hoffmann17). SARS-CoV-2 binds to ACE2 with a receptor-binding domain present at its spike (S) protein (Ref. Reference Hoffmann17). For a successful host cell invasion priming of the S protein by the host proteases such as trans-membrane protease, serine 2 (TMPRSS2), cathepsin-L (CTSL), FURIN or ADAM-17 (a disintegrin and metalloproteinase domain-containing protein 17, also called as TACE) are essential (Refs Reference Hoffmann17, Reference Zipeto18). Considering the essentiality of these host proteins for the entry of the virus inside the host cell, their distribution in the sex-specific tissue may be a deciding factor in creating a sex-specific bias in patient outcomes in COVID-19 (Refs Reference Scully3, Reference Bunders and Altfeld4).
ACE2 and its analogue ‘angiotensin-converting enzyme (ACE)’ are key molecular regulators of renin–angiotensin system (RAS), or renin–angiotensin–aldosterone system, which regulates blood pressure, fluid and electrolyte balance, and systemic vascular resistance. ACE2 competes with its analogue ACE to keep a balance of the pathways regulating these functions (Fig. 2). SARS-CoV-2 can potentially downregulate ACE2 (but not ACE) in the infected epithelium of the lung (Refs Reference Glowacka19, Reference Kumar20). A SARS-CoV-2-induced downregulation of ACE2 in lung epithelium (and perhaps in other tissues including vascular endothelium) can favour the ACE/Ang II/AT1R-mediated pathway leading to vasoconstriction, lung fibrosis and increased inflammation (Ref. Reference Zipeto18).

Fig. 2. Virus-mediated modulation of RAS in COVID-19 patients and influence of sex hormones. ACE2 and its analogue ACE are key molecular regulators of RAS, which regulates blood pressure, fluid and electrolyte balance and systemic vascular resistance. ACE2 competes with its analogue ACE to keep a balance of the pathways regulating these functions. ACE2 metabolises Ang II to Ang 1–7 which further act through Mas1R present in lung epithelial cells favouring vasodilatation, anti-inflammation and antifibrosis. Conversely, an ACE prevents metabolising Ang II which further acts on its cognate receptor AT1R favouring vasoconstriction, fibrosis and increased inflammation. SARS-CoV-2 potentially downregulates ACE2 (but not ACE) in the infected epithelial cells thus favours an ACE/Ang II/AT1R-mediated pathway leading to vasoconstriction, lung fibrosis and increased inflammation resulting in increased disease severity. Sex hormones can effectively modulate RAS axis influencing severity of disease in COVID-19 patients. Higher serum levels of oestrogen in females plausibly downregulates ACE and AT1R, and upregulates ACE2 and Mas1R, thus favours ACE2/Ang 1–7/Mas1R-mediated regulation of RAS resulting in less severe disease. Conversely, higher serum levels of testosterone in male produces an opposite effect. Testosterone by regulating expression of TMPRSS2 gene through AR causes increased internalisation of ACE2: SARS-COV-2 complex. The increased consumption of ACE2 by SARS-CoV-2 causes its depletion on epithelial cells thus activating the ACE/Ang II/AT1R-mediated pathway favouring vasoconstriction, increased tissue inflammation and fibrosis resulting in severe COVID-19. ACE, angiotensin converting enzyme; Ang II, angiotensin II; AT1R, angiotensin type II receptor 1; Mas1R, Mas1 proto-oncogene, G protein-coupled receptor.
Multiple studies have shown a higher expression of ACE2 in men, specifically in reproductive tissues and more particularly in testis (testosterone producing Leydig cells, supportive Sertoli cells and seminiferous tubules which harbour spermatogonia) (Refs Reference Wang and Xu21–Reference Xu23) (Fig. 3). Expression of TMPRSS2 is also significantly higher in reproductive tissues of men (Fig. 3). Accordingly, inflammation, leucocyte infiltration, damage of testicular tissue components and cell death, in post-mortem examinations and complaints of testicular pain in living COVID-19 patients were also reported. Similar evidence was also presented in SARS (Refs Reference Xu23–Reference Cardona Maya, Du Plessis and Velilla25). However, no direct evidence for the presence of SARS-CoV-2 in testis of patients with COVID-19 could be shown yet. Conflicting reports are available for SARS-CoV-1 in human testis (but not for MERS) (Refs Reference Xu23, Reference Zhao26).

Fig. 3. Tissue-specific distribution (m-RNA and protein) of SARS-CoV-2 host cell entry receptor (ACE2) and related proteases (TMPRSS2, FURIN and ADAM17) in reproductive system components in men. A high expression (m-RNA and/or proteomic) of ACE2 and significantly increased expression of the one or more proteases is observable in multiple cellular/tissue components indicating their high susceptibility for the SARS-CoV-2 infection. Data source: Human Protein Atlas (https://www.proteinatlas.org/humanproteome/sars-cov-2, an open access database).
Conversely, expressions of other host proteases, CTSL, FURIN and ADAM17, were significantly higher in women reproductive tissue (Fig. 4) (Ref. 27). Of note, among the host proteases only TMPRSS2 has a known androgenic gene regulation which can influence its tissue expression hence may be a reason for its higher expression in men (Refs Reference Bunders and Altfeld4, Reference Montopoli28). Testosterone by regulating expression of TMPRSS2 gene through androgen receptor (AR) causes increased internalisation of ACE2: SARS-CoV-2 complex (Ref. Reference Mohamed29). The increased consumption of ACE2 by SARS-CoV-2 causes its depletion on lung epithelial cells and other tissue including vascular endothelium, thus activating the ACE/Ang II/AT1R-mediated pathway leading to vasoconstriction, increased tissue inflammation and fibrosis (Fig. 2). However, results from a recent preliminary study (Ref. Reference Baratchian30) discouraged for androgen-mediated TMPRSS2 regulation in COVID-19 outcomes, at least for the pulmonary symptoms. Investigators of this study found no evidence for androgen regulation of pulmonary TMPRSS2 explaining sex-discordant COVID-19 outcomes in human as well as mice. However, they found sex-discordant expressions of AR and ACE2; AR showed limited expression in airway cells of human males but was absent in human females. In mice, males expressed significantly higher ACE2 protein in airways compared with females. They showed in mice, TMPRSS2 expression was unaffected and ACE2 modestly suppressed by potent AR blockade which led them to conclude that that sex differences in COVID-19 outcomes attributable to viral entry are independent of TMPRSS2 however ACE2 may have a role. Their results did not rule out a possible role of TMPRSS2-mediated mechanism in tissues other than lung (Ref. Reference Baratchian30).

Fig. 4. Tissue-specific distribution (m-RNA and protein) of SARS-CoV-2 host cell entry receptor (ACE2) and related proteases (TMPRSS2, FURIN and ADAM17) in reproductive system components in women. A low or non-detectable m-RNA and/or proteomic expression of ACE2, however, significant expression of one or more proteases is observable across the tissues, indicating their low susceptibility for SARS-CoV-2 infection. Data source: Human Protein Atlas (https://www.proteinatlas.org/humanproteome/sars-cov-2, an open access database).
Strong indications for the role of androgenic regulation of ACE2 in COVID-19 vulnerability in men and possible benefits from the use of anti-androgenic drugs were received from a recent study by Samuel et al. (Ref. Reference Samuel31). These investigators applied in vitro high-throughput drug screening to identify drugs that reduce ACE2 levels in human embryonic stem cell (hESC)-derived cardiac cells and lung organoids. Target analysis of hit compounds using bioinformatics revealed androgen signalling as a key modulator of ACE2 levels. Furthermore, they treated hESC-derived lung organoids with anti-androgenic drugs, which reduced ACE2 expression and protected the cells against SARS-CoV-2 infection. Investigators also examined clinical data on COVID-19 patients showing that prostate diseases, which are linked to elevated androgen, are significant risk factors and that the presence of genetic variants in the patients that increase androgen levels is associated with higher disease severity (Ref. Reference Samuel31).
Role of evolution and genetics, epigenetic mechanisms and sex hormones
Evolution and genetics
The good health of females is crucial for population growth of a species, as they raise offspring. Females may pass the protective immune response against the infectious agents to their babies in uterus and through breast milk, thus helping their survival. Stronger immune responses against infections in the case of females have an evolutionary trail which is evident across the species (Refs Reference Úbeda and Jansen5, Reference Zuk, Stoehr, Klein and Roberts6). The underlying mechanisms favouring immunological gain for the females are not well understood; however, genetic landscape of immune genes of the organisms provides some significant clues, as they show a female dominance in their expressions (Ref. Reference Schurz32). First such clue is the existence of dual X-chromosome in females. However, the extra X-chromosome does not remain functional because of a phenomenon, known as X-chromosome inactivation (XCI), in females (Fig. 5). Some of the X-linked genes (approximately 15% of X genes in humans and 3% in mice) in the females escape XCI, providing double dosage of those genes (Refs Reference Schurz32, Reference Klein and Flanagan33).

Fig. 5. Schematic representation of the role of evolution, genetics, epigenetic mechanisms and sex hormones in sex-based differences in COVID-19 outcomes. Females bear two X chromosomes, of which one gets inactivated during oogenesis through epigenetic silencing of the genes – this phenomenon is known as ‘X-chromosomal inactivation’ (XCI). Some of the X-linked genes escape XCI, among these are the key immune functions genes, such as ACE2, TLRs-7, -8, ILs-4, -10 and -13, FoxP3, CD40L, IRAK1 and NEMO, thus these genes express in females in double dosage in comparison with males. These immune genes not only prime the females for a stronger immune response against infectious agents, but also prevent against hyper-inflammatory responses, such as CS. Apart from this, certain immune genes have sex hormone-specific regulatory elements in their promoter region modulating their expression, such as oestrogen has for IFN, and TMPRSS2 has for TMPRSS2. A marked influence of sex hormones on the formation of cytokines has also been noted, such as testosterone induces greater syntheses of Th-17 cells and in turn release of pro-inflammatory markers in males, such as IL-17, IL-20 and CCL-20 – thus favour hyper-inflammatory responses and in turn poor clinical outcomes. XCI, X chromosome inactivation; ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease, serine 2; ARE, androgen receptor element; SRY, sex-determining region Y; SOX-9, SRY-box transcription factor-9; TLRs, Toll-like receptors; IFN, interferon; ILs, interleukins; FoxP3, forkhead box P3; CD40L, cluster of differentiation 40L; CCL-20, chemokine (C–C motif) ligand-20; IRAK1, interleukin-1 receptor-associated kinase 1; NEMO, NF-κB essential modulator.
X-chromosome is known to contain many immune response genes which escape XCI, notably, IL-4, IL-10, IL-13, FoxP3, CD-40L and TLRs-7 and -8 – the pattern recognition receptors – mediating host innate response against RNA viruses, including coronaviruses (Refs Reference Bunders and Altfeld4, Reference Schurz32). IL-4, -10 and -13 are anti-inflammatory ILs (Ref. Reference Kucharzik34) hence their abundance in females in comparison with the males might be a reason for less severe inflammatory disease in them. Interestingly, FoxP3, that is a lineage specification factor for the T-regulatory (T-reg) cells (a type of CD4+ T cells), has oestrogen response elements in the promoter gene region (Ref. Reference Rudensky35). T-reg cells are essential for maintaining immuno-tolerance, preventing autoimmunity and limiting inflammations (Ref. Reference Rudensky35). CD40L is involved in CD4+ T-cell-mediated help for B cells (Ref. Reference Tukiainen36). Thus, the abundance of FoxP3 and CD40L may contribute to the anti-inflammatory control preventing hyper inflammation, and stronger antibody-mediated response in females (Ref. Reference Bunders and Altfeld4).
Apart from these, interleukin 1 receptor associated kinase 1 (IRAK1), a key regulatory molecule in the TLR-dependent signalling pathway, and a number of genes coding for immune regulatory molecules functioning downstream in the cytokine receptor signalling, such as NF-κB essential modulator (NEMO) modulates NF-κB expression, are also X-linked (Ref. Reference Schurz32).
A more robust innate immune response in the early course of COVID-19 may give women a survival advantage over men. Additionally, the gene for the SARS-CoV-2 host cell entry receptor – ACE2 – is located on the X-chromosome, which provides a potential basis for sex-biased expression of this protein. Moreover, ACE2 is an ISG, which is in turn bears oestrogen regulatory elements in its promoter region, which can further explain a favourable innate immune response in females (Refs Reference Scully3, Reference Ziegler37).
Conversely, Y-linked genes, such as Sry (testis-determining factor), Sox9 (SRY-box 9), may be responsible for a dampening influence of testosterone on innate immune response in men (Ref. Reference Klein and Flanagan33). Additionally, TMPRSS2 gene has a testosterone-inducible element in its promoter region, hence, it can be a key influencing factor in men (Refs Reference Scully3, Reference Stopsack38).
Sex linkage of viral cell entry receptor or immune molecule polymorphs might be a reason for higher vulnerability for the infection and increased disease severity in some individuals. Such variants have recently been reported for ACE2 (Refs Reference Benetti39, Reference Cao40) and TLR7 (Ref. Reference van der Made41), both of which are X-linked molecules. In a case series study involving four young male patients, a rare mutation of TLR7 was associated with impaired IFN (types I and II) responses in patients with severe COVID-19 (Ref. Reference van der Made41). Polymorphs have also been reported for TMPRSS2 (Ref. Reference Hou42), which has androgenic regulation at the gene level (Ref. Reference Scully3). However, none of these studies could show a clear causal relationship for the patient outcomes favouring women.
Epigenetics mechanisms
Epigenetics is the study of genetic mechanisms which alter the expression of genes without altering the genetic code of an organism. Salient examples of epigenetic mechanisms are modifications in the chemical tags (such as methyl or hydroxyl groups) of the nucleotide bases in the DNA or modifications in the histone proteins covering the DNA, which silence or activate a gene. Additionally, the post-transcriptional silencing of gene expression through non-coding RNAs, such as microRNAs (miRNAs) and long-noncoding RNAs (lncRNAs), is also considered an epigenetic mechanism. Escape from XCI, influences of sex hormones and immunological tolerance are mediated through epigenetic mechanisms (Fig. 5) (Refs Reference Bunders and Altfeld4, Reference Klein and Flanagan33).
Several miRNA and lncRNA expressions are involved in immune response activation specific to particular sex; particularly, miRNAs which regulated TLR-7 (Ref. Reference Feng43). miRNAs are abundant on the X-chromosome and are known to be modulated by sex hormones. The X chromosome contains 10% of the ~800 miRNAs in the human genome, in comparison, the Y chromosome contains only two miRNAs. miRNAs – including miRNA-18 and miRNA-19, which are encoded on the X chromosome – have known roles in sex differences in immune responses against infectious agents, including viruses (Ref. Reference Klein and Flanagan33). Pontecorvi et al., using a bioinformatics approach, identified a set of miRNAs involved in sex-specific modulation of SARS-CoV-2 host cell entry proteins (Ref. Reference Pontecorvi44). Wet-lab studies investigating the sex-specific role of miRNAs and lncRNAs in COVID-19, are at the infantile stage now and further specific studies will reveal its insight mechanistic details.
Sex hormones
Lymphocytes and other immune cells express sex hormone receptors. Oestrogen response elements are present in majority of immune responsive genes, which facilitate a stronger immune response in adult females. Animal model studies showed that oestrogens promoted the development of antibodies against infectious agents, including respiratory viruses (Refs Reference Klein and Flanagan33, Reference Fink45, Reference Flanagan46). Existing literature suggests that oestrogen plausibly downregulates ACE and angiotensin type II (Ang II) receptor 1 (AT1R), and upregulates expressions of ACE2 and Mas 1 receptors in epithelial cells (REF) thus favours the ACE2/Ang 1-7/Mas1R-mediated pathway leading to vasodilatation, anti-inflammation and antifibrosis (Fig. 2) (Refs Reference Zipeto18, Reference Bukowska47). Another female sex hormone progesterone also has anti-inflammatory and protective effects against viral infections of mucosal epithelium (Refs Reference Viveiros48, Reference Hall and Klein49).
Conversely, androgens, including testosterone and dihydrotestosterone, have an immunosuppressive influence (that extends over pro-inflammatory as well as anti-inflammatory marker genes). Studies showed that testosterone suppressed the development of antibodies in mice (Refs Reference Klein and Flanagan33, Reference Fink45–Reference Gadi51). Lower immune responses to influenza vaccination were reported in men, particularly those with high levels of testosterone (Ref. Reference Furman52).
Testosterone is also linked with higher expression of the Th-17 pro-inflammatory marker in men which is associated with increased disease severity (Refs Reference Bunders and Altfeld4, Reference Klein and Flanagan33, Reference Gadi51). Men with androgen deficiencies have higher concentrations of inflammatory cytokines, such as IL-1β, IL-2 and tumour necrosis factor (TNF), antibody titres and CD4/CD8T cell ratios compared with men with basal testosterone levels (Refs Reference Klein and Flanagan33, Reference Furman52, Reference Mohamad53). Furthermore, men treated with a gonadotropin-releasing hormone (GnRH) antagonist (which significantly reduces testosterone levels), have lower counts of T-reg cells and higher counts of natural killer cells in peripheral blood compared with placebo-treated men or men treated with both, GnRH antagonist and exogenous testosterone (Ref. Reference Klein and Flanagan33).
In a recent prospective cohort study (Ref. Reference Çayan54), Cyan et al. reported significantly increased chance of ICU admissions (P = 0.001) and mortality (P = 0.002) with lower serum total testosterone levels in male patients with COVID-19. Investigators also noted post-infection significant decrease in total serum testosterone levels (from 458 ± 198 to 315 ± 120 ng/dl; P = 0.003). A decrease in the total serum testosterone was significantly correlated with an increase in age (r = 0.236; P = 0.000) and increased levels of pro-inflammatory markers, such as D-dimer (r = 0.213; P = 0.003), procalcitonin (r = 0.225; P = 0.000) and C-reactive protein (CRP) (r = 0.144; P = 0.003), which indicated the role of age and viral-induced pathogenesis in testosterone production in male patients with COVID-19 (Ref. Reference Çayan54).
A higher level of testosterone in male may be activating the ACE/Ang II/AT1R pathway of RAS in lung epithelium (and perhaps also in other tissues including vascular endothelium) leading to an increased severity of COVID-19 (Fig. 2) (Ref. Reference Mohamed29), although any concrete in vivo evidence is currently lacking in this regard.
Numerous studies clearly suggest that sex hormones have an impact on individual's immune response however, that is age-dependent, and above described patterns are mostly limited to reproductive age (Refs Reference Bunders and Altfeld4, Reference Klein and Flanagan33). In the case of children and elderly (>80 years), the influence of sex hormones over immune response is not much distinguishable (Ref. Reference Bhopal and Bhopal55).
Figure 4 presents a summarised account of the role of evolution, genetics, epigenetic mechanisms and sex hormones in sex-based differences in COVID-19 outcomes.
Immunological factors
An increasing body of evidence established immunological factors are key determinants of the disease severity and patient outcomes in COVID-19 (Ref. Reference Kumar56); hence it is likely that they play significant roles in observed sex bias in the presentation of the cases. A hyperactive innate immune response against the invading pathogen leading to systemic hyper-inflammatory state – ‘cytokine storm (CS)’ – is a well-known phenomenon deciding disease severity in respiratory viral infections, such as SARS and influenza (Refs Reference Kumar56, Reference Coperchini57). Clinical studies demonstrated that CS plays a very critical role in developing severe COVID-19 (Refs Reference Kumar56, Reference Coperchini57). CS in COVID-19 is characterised by increased levels of a unique set of cytokines (and chemokines), such as IL-2, IL-6, IL-7, IL-10, granulocyte colony stimulating factor and granulocyte-macrophage colony stimulating factor, interferon-gamma inducible protein 10 (IP10), monocyte chemo-attractant protein 1, macrophage inflammatory protein 1-α, CRP and ferritin, TNFα and IFNγ. Particularly, circulating concentrations of CXCL10, CCL2, IL-2R, IL-6, TNFα, CRP and ferritin are found significantly increased in the patients requiring ICU admission (Refs Reference Kumar56, Reference Coperchini57). IL-6 and IL-10 were reported as the key prognostic markers indicating disease severity in COVID-19 (Refs Reference Dhar58, Reference Zhang59).
As we discussed above (in subsection ‘Evolution and genetics’), a sex-based difference in the immune response can be largely attributed to differential regulation of the protective genes, which seem to be evolutionarily biased in favour of females, supposedly for ensuring better survival of the species (Refs Reference Scully3, Reference Ziegler37). Females not only show a more robust protective immune response but also develop lower tissue injury and have better tissue repair (Refs Reference Fuseini60–Reference Pociask64); these factors taken together can be reason they develop milder disease forms in comparison with males, including lower chances of developing systemic hyper-inflammatory states, such as CS.
In vitro and in vivo studies suggest that females have relatively a higher number of innate immune cells as well as they form stronger innate immune responses (Refs Reference Bunders and Altfeld4, Reference Klein and Flanagan33). A stronger TLR-7-mediated IFN response and higher basal production of IFN and IFN regulatory factor 5 (IRF5) were noted in vitro and animal model studies in the case of female organisms. In humans, IFN type I responses are differentially regulated between men and women (Ref. Reference Ziegler and Altfeld65). Females also exhibited a stronger adaptive (cell and antibody mediated) immune responses in comparison with males. Basal levels of immunoglobulin as well as antibody responses were consistently higher in females compared with those in males (Fig. 6).

Fig. 6. Immunological basis of sex-based differences in COVID-19 outcomes. A stronger protective immune response, for its each component: TLR-based viral sensing, and innate and adaptive immunity, is ensued against SARS-CoV-2 infection in females in comparison with males. TLR, Toll-like receptor; IFN, interferon; ISGs, interferon-stimulated genes; ILs, interleukins; IRF, interferon regulatory factor; CD, cluster of differentiation; RORγt, retinoic acid-related orphan receptor gamma-t; MAVS, mitochondrial antiviral signalling protein; IRAK, interleukin-1 receptor-associated kinase; TRAF, TNF receptor (TNFR) associated factor; TBK, TANK-binding kinase; MyD88, myeloid differentiation primary response gene 88.
The expressions of TLR-pathway and pro-inflammatory genes [e.g. TLR7, myeloid differentiation primary response gene 88 (MYD88), retinoic acid inducible gene-I (RIG-I), IRF7, IFN-β, Janus kinase 2 (JAK2), signal transducer and activator of transcription (STAT3), nuclear factor-κB (NFKB), IFN-γ and tumour necrosis factor (TNF)] were higher in female compared with those in male peripheral blood mononuclear cells from humans and tissues from rats (Ref. Reference Griesbeck66).
Remarkably, the presence of mutated forms of TLR7 was found associated with severe COVID-19 in male patients in a preliminary study (Ref. Reference van der Made41). Coronaviruses, including MERS-CoV and SARS-CoV-1, can induce type I IFN responses through TLR7 (Refs Reference Karnam67–Reference Li69), and SARS-CoV-1 and SARS-CoV-2 have been shown to be sensitive to viral restriction by ISGs in vivo and in vitro (Refs Reference Haagmans70, Reference Mantlo71). Several studies have demonstrated that women produce more IFN type I in response to TLR7 ligands, including viral RNAs, resulting in higher induction of ISGs in women (Refs Reference Berghöfer72–Reference Seillet75). The initial studies assessing type I IFN responses induced by SARS-CoV-2 infections suggested induction of ISGs by SARS-CoV-2 (Refs Reference Blanco-Melo76–Reference Zhou78), but to a lower level than observed in SARS-CoV infection (Refs Reference Blanco-Melo76, Reference Chu77), indicating a potential immune evasion mechanism (Fig. 6) (Ref. Reference Bunders and Altfeld4).
Human studies have shown, regardless of the age, females have not only higher expressions of immunoglobulins and antibodies, but they also have higher expression of immune function genes in the B cells, higher count and activity of helper and cytotoxic T cells and higher CD4+:CD8+ T cell ratio (Refs Reference Scully3, Reference Bunders and Altfeld4, Reference Seillet75). However, as a drawback of stronger immunity, women also have the greater susceptibility to autoimmune diseases (Refs Reference Scully3, Reference Bunders and Altfeld4, Reference Seillet75). Keeping in that line, in patients with COVID-19, a more favourable immune response has been observed in women. Takahashi et al. examined sex differences in viral loads, SARS-CoV-2-specific antibody titres, plasma cytokines and blood cell phenotyping in patients with COVID-19 (Ref. Reference Takahashi1). Investigators found a significant sex-based difference in immune responses. In the patients with moderate disease who had not received immunomodulatory medications, including high dose steroid (prednisolone >40 mg), they observed that men had higher plasma levels of innate immune cytokines, such as IL-8 and IL-18, as well as stronger induction of non-classical monocytes. In contrast, female patients, including the aged, showed significantly stronger T cell activation (Ref. Reference Takahashi1). They additionally found that a poor T cell response negatively correlated with patients' age and led to poor outcome, which was more prominent in men. However, higher innate immune cytokines were associated with worse disease progression in women, but not in men (Ref. Reference Takahashi1).
Some investigators have also shown that SARS-CoV-2 may be detected in nasal swabs for longer periods in men compared with women indicating a delayed viral clearance in men (Refs Reference Zheng79, Reference Xu80).
A faster clearance of the respiratory viral infection in females may be accounted for higher counts of CD4+ Th2 cells and type 2 innate lymphocytes (ILC2s) in them (Ref. Reference Bunders and Altfeld4). Viral clearance from the respiratory mucosa is enhanced by cytokines, such as IL-4, IL-5 and IL-13, which are secreted by type 2 immune cells (Ref. Reference Gause, Rothlin and Loke81). The receptor for IL-13 (IL-13RA1) is encoded by the X chromosome, and may provide women with an advantage of higher gene dosage (Ref. Reference Tukiainen36). Conversely, androgens were shown to reduce ILC2 numbers and cytokine production in male mice (Refs Reference Laffont82, Reference Ricardo-Gonzalez83). Increased expression of IL-13 in females may also protect them through a SARS-CoV-2 host cell entry receptor-based mechanism. IL-13 was found to decrease the expression of ACE2 in epithelial cells in vitro, and respiratory epithelial cells of asthma patients with enhanced type 2 signalling exhibited reduced ACE2 expression compared with healthy controls (Ref. Reference Peters84).
Apart from protection against the infection, immune cell profiles in women are skewed more towards tissue repair responses compared with men, which may have important implications in COVID-19. Women have higher counts of IL-22 producing retinoic acid-related orphan receptor gamma-t (RORγt) + CD4+ T cells which are known to be regulated by sex hormones (Refs Reference Fuseini60–Reference Newcomb62). IL-22 promotes epithelial stem cell proliferation via STAT3 signalling (Ref. Reference Lindemans63), contributing to tissue resilience upon damage. Tissue protective, including lung epithelial repair effects of IL-22, has also been demonstrated in several viral infections (Ref. Reference Pociask64).
How X-linkage of key immune response genes and oestrogenic regulation of IFN can influence the patient's outcomes in infectious diseases has been reflected by a recent COVID-19 study. Bastard et al. examined blood samples from 987 gravely ill COVID-19 patients from multiple ethnic backgrounds across the world. Surprisingly, in at least 101 (10.2%) of the patients who were aged 25–87 years, the investigators found auto-antibodies against IFN type I, of which 95 (94%) were men. However, auto-antibodies were not found in 663 individuals with asymptomatic or mild disease, and were present in only four out of 1227 (0.33%) healthy individuals (Ref. Reference Bastard85).
In addition to the severity of symptoms and mortality, sex-specific immunological responses will have a significant impact on incidence of post-survival health issues in patients with COVID-19, such as functional problems arising from residual organ damages and autoimmune diseases. Possible sex-based variation in the efficacy and adverse effects with vaccines is another crucial indication from the sex-specific immunological responses observed in patients with COVID-19. Sex-specific variations in response to the vaccine were reported in mice models for influenza (however, the same could not be concluded effectively in humans citing data constraints) and it is plausible that it may also hold true in COVID-19 (Refs Reference Fink45, Reference Tadount86).
Figure 6 presents a summarised account of immunological basis of sex-based differences in COVID-19 outcomes.
Additive factors: smoking and gut microbiome
Smoking has been associated with severe pulmonary symptoms and higher mortality in COVID-19 (Refs Reference van Zyl-Smit, Richards and Leone87–89). At the population scale, men are more likely to smoke compared with women, which plausibly can be a contributory reason for the higher overall COVID-19 mortality in men in comparison with women (Ref. Reference van Zyl-Smit, Richards and Leone87). However, in the countries, such as Italy, where there is not wide gender-based difference in the prevalence of smoking, mortality in men remained greater compared with women (Ref. 14) indicating that smoking may have only a limited contribution to sex-specific mortality in COVID-19. Additionally, whether COVID-19 mortality varies between smoking men and women is not sufficiently studied, thus, warrant further study to primarily outline the role of smoking in disease mortality.
Sex-based differences in gut microbiome are considered an important determinant in priming immunological responses against infections, including viruses (Refs Reference Dhar and Mohanty8, Reference Hall and Klein49, Reference Sankaran-Walters90). Recently, it has been noted that sex hormones, specifically oestrogen, have a significant effect on the microbiome of the individuals (Refs Reference Shin91–Reference Baker, Al-Nakkash and Herbst-Kralovetz93). Prominent digestive symptoms are known in patients with COVID-19 and recent studies have shown gut mucosa as a potential site for SARS-CoV-2 infection (Ref. Reference Kumar94). Microbiome is considered to be a crucial factor in the development of gut mucosal immunity against the infections including viruses (Ref. Reference Sankaran-Walters90), hence, may have significant implications for COVID-19 pathogenesis (Ref. Reference Dhar and Mohanty8). The role of gut microbiome in COVID-19-related morbidity and mortality, in reference of sex, has been narrowly studied and warrant further attention.
Non-biological factors
Apart from biological mechanisms, which are the key determinants of the sex-based differences, sex-based differences in COVID-19 mortality can depend on multiple environmental factors, such as gender-based socio-economic disparity and unequal reach to the health-care facilities (Ref. Reference Dehingia and Raj16). An hostile environment added with the health policy discriminations may make a gender more vulnerable for contracting infections and having higher fatality rates, as we have observed a reverse trend in sex-differentiated data for some countries, which showed higher fatality rates in women in comparison with men, which are not being explained biologically (Ref. Reference Dehingia and Raj16).
Concluding remarks
Existing literature and recent research reports provide coherent evidence for the immunological privilege of the women against severe disease symptoms and mortality in COVID-19. This review highlights multiple plausible molecular mechanisms mediating influence of biological sex on mortality in COVID-19 (summarised in Fig. 7 and Table 1), which can be broadly categorised as mechanisms leading to (i) escape of immune response genes present on X chromosome from inactivation, (ii) sex hormones, primarily oestrogen and testosterone, mediated regulation of multiple immune response genes, receptors and tissue proteases and (iii) related to smoking and gut microbiome. An understanding of these molecular mechanisms may help identifying the potential drug targets and developing a sex-differentiated therapeutic approach for COVID-19. Currently, evidence is limited and nascent; further in-depth research will be necessary to obtain a concrete understanding of this issue.
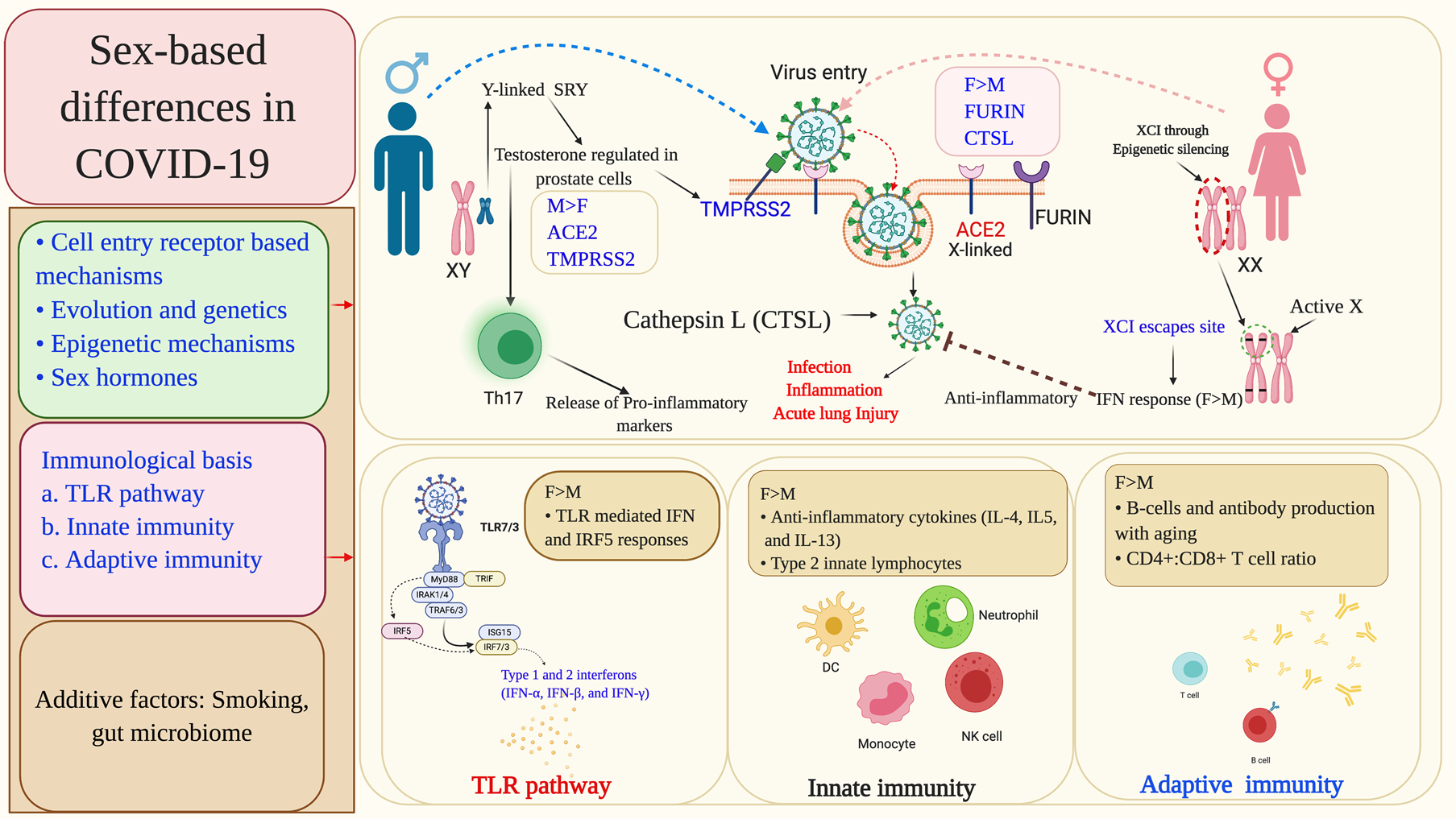
Fig. 7. Summary of the molecular mechanisms leading to sex-differentiated outcomes in COVID-19 patients. The sex-based differences primarily occur at the level of viral cell entry receptors and immune response mechanisms against the infection. Sex-specific genetic and epigenetic factors, and also sex hormones differentially modulate viral cell entry receptors and associated host proteases, viral sensing receptors and multiple immune response genes including IFN. Some other factors, such as smoking and gut microbiome have also been found to influence the patient outcomes in a sex-based manner. XCI, X chromosome inactivation; ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease, serine 2; CTSL, cathepsin L; ARE, androgen receptor element; SRY, sex-determining region Y; SOX-9, SRY-box transcription factor-9; TLRs, Toll-like receptors; IFN, interferon; IRAK1, interleukin-1 receptor-associated kinase 1; ISGs, interferon-stimulated genes; IRF-interferon regulatory factor; TRAF, TNF receptor (TNFR) associated factor; TRIF, TIR-domain-containing adapter-inducing interferon-β; MyD88, myeloid differentiation primary response gene 88.
Table 1. Molecular mechanisms for sex-based differences in patient outcomes in COVID-19

Limitations
Our projections for possible molecular mechanisms explaining sex-based difference in patient outcomes in COVID-19 have multiple limitations which need to be considered. Most importantly, currently, original studies in COVID-19, addressing this aspect, are very limited, and available literature largely present speculations based on results from other respiratory viral infections. However, these are important projections which may hold true in COVID-19 if tested, hence have extreme importance for designing future studies. Second, most of the epidemiological and clinical studies involving COVID-19 patients, included in this review, did not ensure standardised control of the confounding factors, such as sex-based disproportions in the included population and sex-biased reach to the medical facilities, which could have affected proportion of the sex-based presentations. However, the general observation across the studies that women with COVID-19 face lower morbidity and mortality is in line with the trend reflected from the global data. Third, there remains possibility that our used key words could not retrieve all studies under targeted categories, especially, many recent preliminary studies or preprint articles, which may have relevant findings.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/erm.2021.9
Acknowledgement
We acknowledge help of databases, Human Protein Atlas (https://www.proteinatlas.org/) and Global health 50/50 (https://globalhealth5050.org) for making available COVID-19-related data used in this study.
Author contributions
A.K. wrote the first draft. R.K.N., M.K., P.P., C.K., V.P., S.K., P. S. S., K.S., K.K., and S. K. revised the draft. R.K.N. performed data analysis, and P.P. and R.K.N. prepared the figures.
Conflict of interest
The authors declared ‘no conflicts of interest’.










