Introduction
Until the 1950s, farmers used organic and inorganic fertilisers in soil management to improve plant growth (Novotny, Reference Notvony1999). Thereafter, the growing world population encouraged the development and application of new technologies and intensified management practices to increase production per cultivated area (Stewart et al., Reference Stewart, Dibb, Johnston and Smyth2005). Thus, the use of synthetic fertilisers in agricultural systems increased food production by about 50% (Skowrońska and Filipek, Reference Skowronska and Filipek2014), supporting population growth and economic development (Zhang et al., Reference Zhang, Davidson, Mauzerall, Searchinger, Dumas and Shen2015). However, some studies have shown that the amounts of synthetic fertilisers applied to the soil increased almost exponentially (Skowrońska and Filipek, Reference Skowronska and Filipek2014). In addition to the increasing production costs, such practice causes negative environmental effects (Mondal et al., Reference Mondal, Datta and Mondal2017), including eutrophication of water bodies, groundwater contamination, air pollution and soil impoverishment, mainly due to the application of synthetic nitrogen (N) fertilisers to the soil (Vitousek et al., Reference Vitousek, Aber, Howarth, Likens, Matson, Schindler, Schlesinger and Tilman1997). Therefore, looking for clean and sustainable alternatives is a challenge for modern sustainable agriculture.
The inoculation of plant growth-promoting bacteria is a reasonable alternative to the use of synthetic fertilisers, especially those nitrogenate (Vargas et al., Reference Vargas, Lisboa, Schlindwein, Granada, Giongo, Beneduzi and Passaglia2009). Such bacteria group can promote plant growth through biological nitrogen fixation (BNF), solubilisation of nutrients such as phosphorous (P) and potassium (K), hormone production and inhibition of diseases caused by soil-borne pathogens (Schmidt et al., Reference Schmidt, Thomé, Sperotto and Granada2019; Volpiano et al., Reference Volpiano, Lisboa, São José, Oliveira, Beneduzi, Passaglia and Vargas2018). Legumes can benefit from the inoculation of symbiotic N2-fixing bacteria, which may be sufficient to supply all the N needs for crop development and to promote plant growth with acceptable yield/productivity (Gourion et al., Reference Gourion, Berrabah, Ratet and Stacey2015). This inoculation also improves soil fertility (Bhattacharyya and Jha, Reference Bhattacharyya and Jha2012), reducing the need of synthetic N fertilisers for subsequent crops. Therefore, selection of rhizobia isolates for legume production is an important approach to achieve environmental and agricultural sustainability (Sengupta and Gunri, Reference Sengupta and Gunri2015).
Common beans (Phaseolus vulgaris L.) are one of the most important grains for human consumption, showing great economic, nutritional and cultural relevance for several countries (Yanni et al., Reference Yanni, Zidan, Dazzo, Rizk, Mehesen, Abdelfattah and Elsadany2016). In 2016, the estimated global production of dry beans was approximately 27 million tons. According to FAO (2018), Asia accounts for 45.6% of total production, while the Americas produce 26.1%, followed by Africa (24.2%), Europe (4.0%) and Oceania (0.1%). Common bean plants benefit from BNF, although frequently with low efficiency. Such low BNF efficiency can be attributed to short crop cycle and promiscuous establishment of symbiosis with a wide variety of native rhizobial species (Shamseldin and Velázquez, Reference Shamseldin and Velázquez2020). The rhizobial inoculation, especially indigenous strains adapted to the local edaphoclimatic conditions, is proven to be an effective strategy to overcome these problems (Pastor-Bueis et al., Reference Pastor-Bueis, Sánchez-Cañizares, James and González-Andrés2019). In this context, some recent studies have shown that inoculation of new rhizobial isolates in common bean plants can eliminate the need of N fertilisation under greenhouse conditions (Stajković et al., Reference Stajković, Delić, Josić, Kuzmanović, Rasulić and Knezević-Vukčević2011; de Souza et al., Reference de Souza, Bassani, Sperotto and Granada2016; Fageria et al., Reference Fageria, Melo, Ferreira, Oliveira and Knupp2014). However, field experiments and the recommendation of new rhizobia isolates are gaps in the knowledge transference from laboratories to producers.
Selection of new indigenous rhizobial strains for inoculation in leguminous and their testing under field conditions to reduce the use of synthetic N fertilisers in the soil are relevant biotechnological tools. Thus, this study evaluated the agronomic attributes of common bean plants cultivated under field conditions and inoculated with two rhizobial strains (previously isolated from common bean roots – de Souza et al., Reference de Souza, Bassani, Sperotto and Granada2016).
Material and Methods
Organisms and 16S rRNA sequence analysis
Bacteria SEMIA 4108 and SEMIA 4107 were used in this study. These strains were previously isolated by de Souza et al. (Reference de Souza, Bassani, Sperotto and Granada2016) as VC28 and M3, respectively, and are currently available at the ‘SEMIA culture collection’ (IBP World Catalogue of Rhizobium Collections no. 443 in the WFCC World Data Centre on Microorganisms). To further characterise both strains, the genomic DNA was isolated from bacterial cells using the PureLink™ Genomic DNA Mini Kit (Thermo Scientific). The near-full length 16S rRNA gene was amplified using the primers BacPaeF (5ʼAGAGTTTGATCCTGGCTCAG3’) and Bac1542R (5’AGAAAGGAGGTGATCCAGCC3’), according to Granada et al. (Reference Granada, Beneduzi, Lisboa, Turchetto-Zolet, Vargas and Passaglia2015). Nucleotide sequences were sequenced on both strands of PCR amplification products using the Macrogen sequencing equipment (Macrogen Inc., Seoul, South Korea), an ABI3730XL and sequencing primers 785F and 907R. Low-quality sequences were trimmed using the Chromas 2.6.4 software. Fragments were assembled into a single 1,450 bp sequence using the EMBOSS merger tool (http://www.bioinformatics.nl/cgi-bin/emboss/merger). Sequence identity was assessed through comparison of 16S rRNA sequences of SEMIA 4108 and SEMIA 4107 isolates with sequences from the EzBioCloud 16S rRNA server database (https://www.ezbiocloud.net/identify). The sequence data reported in this study are publicly deposited at the GenBank under the accession numbers MN209793 and MN209795.
A 16S rRNA phylogeny was reconstructed with Rhizobium isolates SEMIA 4108 and SEMIA 4107, 90 strains of Rhizobium spp. and Ensifer adhaerens ATCC 33212 as outgroup, according to the sequence accessions for taxonomic studies provided on LPSN (available at https://lpsn.dsmz.de/). The 16S rRNA sequences were aligned with the AlignSeqs function available in ‘DECIPHER’ v2.14.0 R package (Wright, Reference Wright2015). The ‘phangorn’ v2.5.5 R package (Schliep, Reference Schliep2011) was used to select generalised time-reversible with gamma rate variation (GTR+G+I) as the best-fitting model, according to the Akaike information criterion using ModelTest function. Then, ‘phangorn’ functions were also used to construct phylogenetic trees using the maximum likelihood (ML) estimation. We first constructed a neighbour-joining tree, and then fit the GTR+G+I ML tree using the neighbour-joining tree as a starting point. A protocol with an explicit number of 500 bootstraps was processed. The trees were visualised and edited using the R packages phytools (Revell, Reference Revell2012) and ggtree (Yu et al., Reference Yu, Smith, Zhu, Guan and Lam2017).
Field experiments
Field experiments were carried out at the Viamão Research Centre (30°2ʼ10.64"S and 51°1ʼ17.65"W, 71 m a.s.l.) of the Department of Agricultural Research and Diagnosis of the State of Rio Grande do Sul, located in Viamão RS, Brazil. The soil of this region is classified as Ultisol (Soil Survey Staff, 1999) and, according to the Köppen’s classification, the regional climate is subtropical with warm and wet summers (Cfa). Climate records were obtained from a local weather station, providing information on minimum and maximum air temperature, relative humidity and rainfall during experiments. To evaluate soil fertility, soil nutrient analyses were performed before soil corrections. Organic matter (OM), clay, macro and micronutrients and pH were determined using the standard methods described by Sparks et al. (Reference Sparks, Page, Helmke and Loeppert2020). Field experiment was performed according to the Official Protocol for Evaluation of Agronomic Efficiency of Inoculants related to BNF processes in legumes (Brazilian Ministry of Agriculture, Livestock, and Supply – MAPA – BRASIL, 2011) during the growing periods of 2016/17 and 2018/19 (October to January).
Common bean seeds were inoculated with rhizobial isolates previously described by de Souza et al. (Reference de Souza, Bassani, Sperotto and Granada2016). SEMIA 4108 was characterised as a strong siderophore and low indolic compounds producer, while SEMIA 4107 was characterised as a strong siderophore and high indolic compounds producer. Additionally, a treatment with SEMIA 4088 (Rhizobium tropici), a rhizobial strain approved by MAPA as an inoculant for common bean cultivation, was performed.
For the formulation of inocula, rhizobial cells were grown in Yeast Mannitol broth (Vincent, Reference Vincent1970) until reach log-phase. Then, cells were inoculated in sterile peat at a 3:2 (w:v) and stored at room temperature for 10 days. Inoculant quality was monitored on the day before seed inoculation by counting the colony-forming units (CFU) with a minimum viable cell number of 109 CFU per gram of peat. To each kg of common bean seeds (Phaseolus vulgaris L. – Triunfo cultivar), 54 g of the respective inoculant was added.
The experimental design was in randomised blocks with five treatments and four replicates. The area selected for the experiment had never been cultivated with rhizobia inoculation. The five treatments used were (1) negative control (N−), no mineral N fertilisation and without inoculation of rhizobial isolates; (2) positive control (N+), 177 kg ha−1 of urea (recommended dose for common bean plants), with 45 kg at sowing and 132 kg after 20 days of plant emergence and without inoculation of rhizobial isolates; (3) SEMIA 4088, no mineral N fertilisation and inoculation with recommended rhizobial isolate SEMIA 4088; (4) SEMIA 4108, no mineral N fertilisation and inoculation with rhizobial isolate SEMIA 4108; and (5) SEMIA 4107, no mineral N fertilisation and inoculation with rhizobial isolate SEMIA 4107. All treatments were fertilised with P, K and limestone according to the soil nutrient analysis: 61 and 73 kg ha−1 of triple superphosphate in 2016/17 and 2018/19, respectively; 52 and 69 kg ha−1 of potassium chloride in 2016/17 and 2018/19, respectively. Limestone fertilisation was 2.7 tons ha−1 only in the first experiment. Experimental units had 10 m2 with four rows per experimental unit (0.5 m spaced; Supplementary Figure S1). After seed inoculation, sowing was performed at a density of 14 seeds per meter.
When plants achieved full flowering (R6 stage of growth cycle – around 30 days after plant emergence), ten plants of each experimental unit were harvested, and roots and shoots were separated. Roots were used to determine nodule number and dry matter. Shoots were dried at 65°C for 3 days and used to determine dry matter and N, P and K contents (Sparks et al., Reference Sparks, Page, Helmke and Loeppert2020). At harvest (R9 stage of growth cycle – around 90 days after plant emergence), ten plants were harvested in one linear meter of the two central lines of each experimental unit and used to determine seed yield and N, P and K contents (Sparks et al., Reference Sparks, Page, Helmke and Loeppert2020).
The results obtained were subjected to one-way ANOVA followed by Tukey test (p < 0.05) using the software InfoStat 2018 (Agricultural College of the National University of Córdoba, Argentine). Correlation analyses between plant agronomic attributes observed in both sampling periods (R6 and R9 stages of growth cycle) and growing periods (2016/17 and 2018/19) were performed by principal component analysis.
Results
16S rRNA phylogeny
According to similarity-based searches in databases of 16S rRNA sequences, SEMIA 4108 and SEMIA 4107 had great gene similarity with species belonging to the Rhizobium genus (Supplementary Figure S2). The 16S rRNA phylogeny showed that both SEMIA 4108 and SEMIA 4107 locate next to Rhizobium phaseoli, inside a clade also containing the strains R. acidisoli, R. aegyptiacum, R. aethiopicum, R. anhuiense, R. bangladeshense, R. binae, R. chutanense, R. ecuadorense, R. esperanzae, R. etli, R. fabae, R. indigoferae, R. laguerreae, R. leguminosarum, R. lentis, R. pisi, R. sophorae and R. sophoriradicis (Figure 1).

Figure 1. Cladogram evidencing the clade containing SEMIA 4107 (M3) and 4108 (VC28) strains, inferred by maximum likelihood (ML) analysis. The black circles at the branching points indicate bootstrap values >70%. GenBank IDs are shown after the bacterial species names. Complete phylogeny of 16S rRNA gene sequences from Rhizobium sp. is found in Supplementary Figure S2.
Field experiments
In the experimental field, air temperature varied from 15.8°C to 31.3°C with total rainfall of 556.4 mm in 2016/17, and from 9.0°C to 39.7°C and 395.8 mm in 2018/19 (Supplementary Table S1). The soil was acidic with a medium clay content and low percentage of OM in both growing periods (Table 1). The effective cation exchange capacity (CEC) indicated high contents of P and K (Mehlich-1) and low contents of Ca and Mg, being presented at intermediate levels. The comparison of soil nutrient content in both growing periods showed that 2018/19 presented higher CEC and Ca, and lower Al, P and K than 2016/17 (Table 1). Micronutrients (Cu, Zn and Mn) were available in adequate amounts for common bean cultivation in both periods.
Table 1. Chemical characteristics of soil before amendments

OM, organic matter; CEC, cation exchange capacity.
Agronomic attributes evaluated at the R6 stage (Table 2) showed that plants inoculated with SEMIA 4088 and SEMIA 4108 had higher nodule number compared to that of non-inoculated plants in both growing periods. During 2016/17, plants that received synthetic N fertiliser (N+) had higher shoot dry matter, followed by all inoculated plants, and lastly, the control without inoculation and N fertilisation (N−). Shoot N, P and K concentrations presented the highest values in N+ and the lowest ones in N−. In 2018/19, the shoot dry matter and P content of rhizobia-inoculated plants and N+ control were similar. For N content, N− treatment caused the lowest values, while N+ and SEMIA 4088 induced the highest values, and SEMIA 4108 and 4107 intermediate levels (Table 2).
Table 2. Agronomic attributes of common bean plants evaluated after 30 days of cultivation (R6 stage)

Values followed by the same letter in the same column were not significantly different by the Tukey test (p < 0.05).
At harvest (R9 growth stage), plants inoculated with SEMIA 4107 showed similar productivity to N+ treatment in 2016/17 (Figure 2A) and higher productivity than N+ treatment in 2018/19 (Figure 2E). Considering the amount of nutrients in grains in 2016/17, N accumulation increased from 50 kg ha−1 in N− to more than 60 kg ha−1 in SEMIA 4107 and N+ (Figure 2B), while total K accumulation increased from 26 kg ha−1 in N− to 30 kg ha−1 in SEMIA 4107 (Figure 2D). In 2018/19, N accumulation increased from 31 kg ha−1 in N− to 75 kg ha−1 in plants inoculated with SEMIA 4108 (Figure 2F). Total P accumulation increased from 6.7 kg ha−1 in N− to 15 kg ha−1 in SEMIA 4107 (Figure 2G), and K accumulation increased from 21 kg ha−1 in N− to 52 kg ha−1 in SEMIA 4107 (Figure 2H).
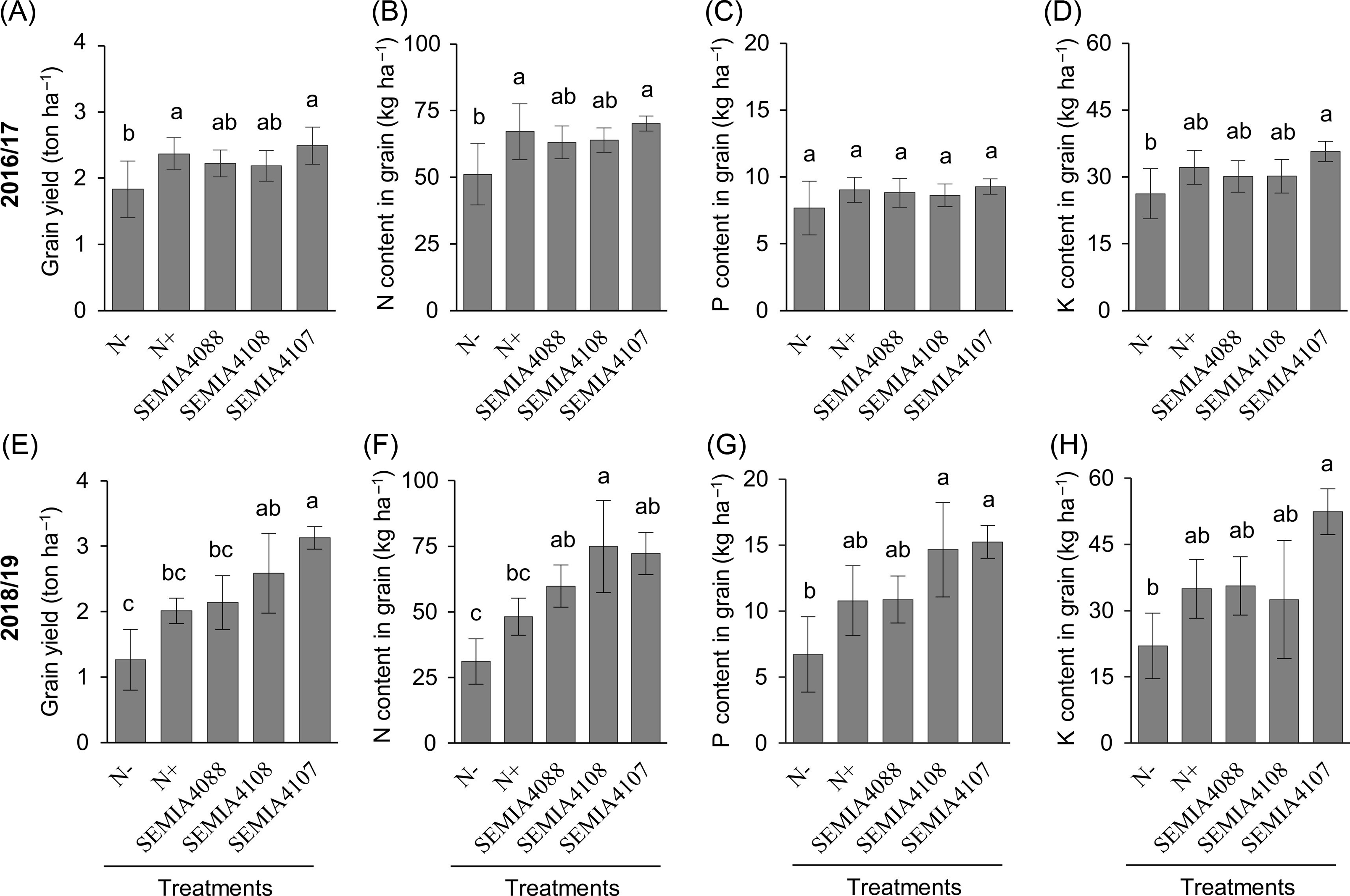
Figure 2. Grain yield and N, P, and K contents of seeds from common bean plants inoculated with rhizobial isolates SEMIA 4088, SEMIA 4108 and SEMIA 4107, and control treatments which received (N+) or not (N−) N fertilisation in 2016/17 (A, B, C, D) and 2018/19 (E, F, G, H). Grain productivity (A, E), grain nitrogen (B, F), phosphorous (C, G) and potassium (D, H) contents. Different letters represent statistical difference between treatments (Tukey test, p < 0.05).
Correlation analysis between agronomic attributes during the R6 and R9 stages of both growing periods helps to better understand our results (Figure 3). The first two principal components (PC1 and PC2) explained 80.2% of the total variability at the first sampling period (R6 stage). Considering all agronomic attributes analysed, inoculated plants (SEMIA 4088, SEMIA 4108 and SEMIA 4107) had correlated results and formed a group, while control treatments (N+ and N−) were arranged out of this group (Figure 3A). Rhizobia-inoculated plants were related to high nodule number and dry matter shoot P content, while N+ was related to high shoot dry matter and shoot N and K contents. At the harvest period (R9 stage), PC1 and PC2 explained 94.2% of the total variability (Figure 3B). The results for plants inoculated with SEMIA 4088, SEMIA 4108 and N+ control were related (forming a group), while SEMIA 4107 was arranged out of this group, showing the most promising results. These data revealed that the development of inoculated plants does not follow the same pattern as N-fertilised plants (N+). Additionally, comparing inoculated treatments with N-fertilised control, no yield or productivity losses at the end of the growing process were detected. The agronomic attributes were similar or even greater than those found in N+ control.

Figure 3. Principal component analysis (PCA) of agronomic attributes of common bean plants inoculated with rhizobial isolates SEMIA 4088, SEMIA 4108 and SEMIA 4107, and control treatments which received (N+) or not (N−) N fertilisation. (A) PCA of attributes evaluated at flowering stage and (B) at harvest, in both 2016/17 (black dots) and 2018/19 (grey dots). PC1 and PC2 account for approximately 80.2% of total variability in (A) and 94.2% in (B).
Discussion
The strains SEMIA 4108 and SEMIA 4107 were previously identified as Rhizobium fabae (de Souza et al., Reference de Souza, Bassani, Sperotto and Granada2016) and a longer 16S rRNA sequence (1,450 bp) analysis was performed to confirm such bacterial identification. To provide reliable identification, as it may be impaired by the quality of sequences deposited in public databases (i.e., NCBI), the curated EzBioCloud 16S rRNA database (Yoon et al., Reference Yoon, Ha, Kwon, Lim, Kim, Seo and Chun2017) was chosen for comparisons. In the phylogenetic tree with 90 Rhizobium strain sequences, SEMIA 4108 and SEMIA 4107 were next to R. phaseoli, inside a clade containing other 18 Rhizobium type strains (Figure 1). This clade contained well-known common bean-nodulating strains, such as R. acidisoli, R. chutanense, R. ecuadorense, R. esperanzae, R. etli and R. leguminosarum. Here, we cannot define our strains at the species level only with 16S rRNA gene analysis. However, the 16S rRNA gene is generally accepted as a standard for identification and classification of prokaryotic species due to its essential function, ubiquity and evolutionary properties (Kolbert and Persing, Reference Kolbert and Persing1999). In 1991, 16S rRNA was incorporated into the minimal standards for Rhizobium species description (Graham et al., Reference Graham, Sadowsky, Keyser, Barnet, Bradley, Cooper and Young1991) and the exact copy number and variations in 16S rRNA sequences can be easily determined. The limited resolving power of 16S rRNA gene sequences to obtain species identification has become obvious, especially when closely related organisms are being analysed (Hahn et al., Reference Hahn, Jezberová, Koll, Saueressig-Beck and Schmidt2016). Thus, to define their precise species, whole genomes of SEMIA 4108 and SEMIA 4107 are currently being sequenced.
Inoculation of legumes with efficient rhizobial isolates can supply all the N needed for plant development (Peoples et al., Reference Peoples, Brockwell, Herridge, Rochester, Alves, Urquiaga, Boddey, Dakora, Bhattarai, Maskey, Sampet, Rerkasem, Khan, Hauggaard-Nielsen and Jensen2009). Therefore, selection of rhizobia strains for inoculation in different leguminous species aiming at replacing synthetic N fertilisation is a potential biotechnological tool (Koskey et al., Reference Koskey, Mburu, Njeru, Kimiti, Ombori and Maingi2017). The most successful case is Bradyrhizobium spp. inoculation in soybean plants. Strains of Bradyrhizobium japonicum, B. diazoefficiens and B. elkanii are commercialised as specific inoculants for soybean plants around the world. Such inoculation improves soybean growth and productivity without using synthetic N fertilisation (Chibeba et al., Reference Chibeba, Kyei-Boahen, Guimarães, Nogueira and Hungria2018). On the other hand, the knowledge regarding the symbiosis of Rhizobium spp. with common bean plants is still incipient. The promiscuity and fast-growing cycle of common bean plants may reduce BNF efficiency (Kaschuk et al., Reference Kaschuk, Hungria, Andrade and Campo2006; Martínez-Romero, Reference Martínez-Romero2003). However, some studies under greenhouse conditions showed that symbiosis among rhizobia and common bean plants present high BNF potential. Korir et al. (Reference Korir, Mungai, Thuita, Hamba and Masso2017) demonstrated that inoculation of rhizobial strains CIAT 899 and IITA-PAU987 in common bean generates plants with similar shoot dry weight to that of plants receiving N fertilisation. Samavat et al. (Reference Samavat, Mafakheri and Shakouri2012) also reported five rhizobial isolates that improved the growth and nutrient absorption of common bean plants, and the growth-promoting effect was higher when plants were co-inoculated with Pseudomonas fluorescens UTPF68 or UTPF109.
Field experiments with results similar to the ones presented in this work have already been reported. Common bean plants inoculated with CIAT 899 presented approximately 6% higher grain yield than non-inoculated plants (Vargas et al., Reference Vargas, Mendes and Hungria2000), and similar grain yield to N-fertilised plants in Brazilian oxisols (Hungria et al., Reference Hungria, Campo and Mendes2003). These findings agree with our results and confirm the high efficiency obtained by inoculation of common bean plants with selected rhizobial strains. However, we must be aware that soil characteristics and plant cultivars can severely influence inoculation efficiency and plant productivity (Hungria et al., Reference Hungria, Campo and Mendes2003; Zaman-Allah et al., Reference Zaman-Allah, Sifi, L’Taief, El Aouni and Drevon2007).
Herrige et al. (Reference Figueiredo, Martinez, Burity and Chanway2008) estimated that BNF contribution to crop legumes reaches 20–22 million tons of N per year. Due to the losses observed during the application of synthetic N fertilisers, such as urea, it would be necessary to apply approximately 100 million tons of this fertiliser to legume crops to achieve the same N amount provided by BNF (Peoples et al., Reference Peoples, Brockwell, Herridge, Rochester, Alves, Urquiaga, Boddey, Dakora, Bhattarai, Maskey, Sampet, Rerkasem, Khan, Hauggaard-Nielsen and Jensen2009). Considering common bean plants studied in this work, we applied 80 kg N ha−1 in N-fertilised control (N+, 177 kg urea ha−1), and approximately 63 and 50 kg ha−1 of this N were transferred to seeds in 2016/17 and 2018/19, respectively (Figure 2B and F). Plants inoculated with SEMIA 4107 and SEMIA 4108 did not receive N fertilisation, and BNF was able to transfer approximately 70–75 kg ha−1 of N in both growing periods. Thus, besides an environmentally friendly technique, the rhizobial inoculation can provide economic advantages, since application cost of urea is around US$ 93 per hectare, while the inoculant costs around US$ 1.89 per hectare (Mercante et al., Reference Mercante, Otsubo and Brito2017).
Even so, there is a great difficulty in transposing the results obtained in the laboratory and under greenhouse conditions to the field, which consequently hinders the development of new inoculant formulations. In our previous study (de Souza et al., Reference de Souza, Bassani, Sperotto and Granada2016), we performed an initial screening by evaluating the growth-promoting traits of 200 rhizobial isolates. In a greenhouse experiment, common bean plants inoculated with the isolates M3 (SEMIA 4107) and VC28 (SEMIA 4108) showed the greatest BNF and plant growth-promoting potential. A similar screening was performed by Figueiredo et al. (Reference Figueiredo, Martinez, Burity and Chanway2008), Stajković et al. (Reference Stajković, Delić, Josić, Kuzmanović, Rasulić and Knezević-Vukčević2011), Oliveira-Longatti et al. (Reference Oliveira-Longatti, Marra and Moreira2013) and Ribeiro et al. (Reference Ribeiro, Ormeño-Orrillo, Dall’Agnol, Graham, Martinez-Romero and Hungria2013), who found rhizobial isolates with potential for recommendation and commercialisation. However, there are no further reports regarding the continuity of such studies. Herein, we confirm the BNF potential of SEMIA 4107 and SEMIA 4108 in field experiments.
We recognise the importance of synthetic fertilisers for food production, which supported population growth in the last century. However, the use of synthetic N fertilisation in legumes should be revisited and maybe replaced by rhizobial inoculation, which would improve the BNF process and reduce environmental and economic costs. The high biotechnological potential presented by selection and characterisation of new rhizobial isolates for common bean inoculation needs to be deeply investigated.
Conclusion
In this research, we evaluated two promising rhizobial strains (SEMIA 4108 and SEMIA 4107) under field conditions. Sequencing of 16S rRNA showed that both strains are closely related to Rhizobium phaseoli. Common bean plants inoculated with SEMIA 4107 showed higher grain yield compared to the N-fertilised control (N+) and the recommended SEMIA 4088 strain. This agronomical performance confirms that both strains are effective and can sustain common bean growth without nitrogen fertilisation.
Supplementary material
For supplementary material for this article, please visit https://doi.org/10.1017/S0014479721000041
Acknowledgements
We thank the University of Taquari Valley – Univates for the financial support, and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Capes/Brazil) for granting the scholarships.
Disclosure statement
The authors declare no potential conflicts of interest.
Financial support
None.








