Substance misuse is common and associated with widespread consequences at individual and societal levels. Among individuals aged 18–25 years, 57% report drinking alcohol in the past month, and about half of all young adults report use of an illicit substance (Ahrnsbrak, Bose, Hedden, Lipari, & Park-Lee, Reference Ahrnsbrak, Bose, Hedden, Lipari and Park-Lee2017; Schulenberg et al., Reference Schulenberg, Johnston, O'Malley, Bachman, Miech and Patrick2018). Substance use disorders (SUDs) affect approximately 15% of individuals age 12 and older (Ahrnsbrak et al., Reference Ahrnsbrak, Bose, Hedden, Lipari and Park-Lee2017). Risky substance use is associated with immediate and long-term consequences to the individual and the larger community, including physical and mental health challenges, decreased academic performance, relationship problems, crime, and lost wages (Arria et al., Reference Arria, Garnier-Dykstra, Caldeira, Vincent, Winick and O'Grady2013; Gryczynski et al., Reference Gryczynski, Schwartz, O'Grady, Restivo, Mitchell and Jaffe2016; Henkel, Reference Henkel2011; Hingson, Zha, & Weitzman, Reference Hingson, Zha and Weitzman2009; Juibari et al., Reference Juibari, Behrouz, Attaie, Farnia, Golshani, Moradi and Alikhani2018). The financial toll associated with substance abuse is approximately $400 billion annually in the US (National Drug Intelligence Center, 2011; Sacks, Gonzales, Bouchery, Tomedi, & Brewer, Reference Sacks, Gonzales, Bouchery, Tomedi and Brewer2015). Thus, a key focus of substance use research is the optimization of prevention and intervention programs so as to minimize impact of substance-related harms at the individual and societal level.
Current evidence-based approaches to prevent substance-related harms typically involve school-based, multicomponent or family-systems interventions for adolescents, and brief motivational interventions for young adults (Smit, Verdurmen, Monshouwer, & Smit, Reference Smit, Verdurmen, Monshouwer and Smit2008; Spoth, Greenberg, & Turrisi, Reference Spoth, Greenberg and Turrisi2008; Tanner-Smith & Risser, Reference Tanner-Smith and Risser2016). Substantial resources have been dedicated to the design, implementation, and evaluation of such prevention programs, yet most yield small effect sizes (Huh et al., Reference Huh, Mun, Larimer, White, Ray, Rhew and Atkins2015; Sandler et al., Reference Sandler, Wolchik, Cruden, Mahrer, Ahn, Brincks and Brown2014). Furthermore, only about a third of individuals who are treated for SUDs are abstinent immediately posttreatment, and relapse rates remain high (Dutra et al., Reference Dutra, Stathopoulou, Basden, Leyro, Powers and Otto2008; Maisto, Pollock, Cornelius, Lynch, & Martin, Reference Maisto, Pollock, Cornelius, Lynch and Martin2003; Witkiewitz & Masyn, Reference Witkiewitz and Masyn2008).
Why do current prevention and intervention programs continue to yield small effects despite substantial investment? Why do these programs seemingly work for some individuals and not others? Much of the work to understand these limited effects has focused on program implementation and fidelity. There has been less consideration of individual, person-level factors that may differentially influence outcomes (Belsky & van Ijzendoorn, Reference Belsky and van Ijzendoorn2015). However, increasingly, attention has turned to the possible role of genetics in differential response to prevention and intervention.
Genetics has long been known to play an important role in the development of alcohol and substance use problems. Twin studies, which compare concordance rates for a given outcome between monozygotic twins (who share all of their genetic variance) and dizygotic twins (who share on average 50% of their genetic variance), demonstrate that the development of SUDs is partly due to genetics (Tawa, Hall, & Lohoff, Reference Tawa, Hall and Lohoff2016). SUDs are approximately 50–70% heritable, meaning at least half of the variability in liability to SUDs is due to genetic factors (Agrawal & Lynskey, Reference Agrawal and Lynskey2008).
However, genes are only part of the story. Genetic predispositions are known to dynamically interact with environmental factors to contribute to the development of behavioral outcomes (Kendler, Jaffee, & Romer, Reference Kendler, Jaffee and Romer2011). Studies of Gene × Environment (G×E) interaction consistently demonstrate that the importance of genetic effects varies as a function of the environment (Dick & Kendler, Reference Dick and Kendler2012), such that genetic predispositions may be more or less strongly associated with an outcome under certain environmental contexts. For example, an individual who is genetically at risk for alcohol use disorder (AUD) will never develop an AUD if he/she is not exposed to alcohol. The importance of genetic influences on substance use outcomes is known to vary as a function of several environmental factors, including parental monitoring, peer deviance, neighborhood characteristics, and romantic relationship status (Barr et al., Reference Barr, Kuo, Aliev, Latvala, Viken, Rose and Dick2019; Dick et al., Reference Dick, Pagan, Viken, Purcell, Kaprio, Pulkkinen and Rose2007; Harden, Hill, Turkheimer, & Emery, Reference Harden, Hill, Turkheimer and Emery2008; Rose, Dick, Viken, & Kaprio, Reference Rose, Dick, Viken and Kaprio2001; Slutske, Deutsch, & Piasecki, Reference Slutske, Deutsch and Piasecki2019).
Prevention scientists have also studied whether the association between genotype and outcome varies as a function of participating in a prevention or intervention program. In this way, intervention status is the environment that moderates genotype-outcome associations. Gene-by-intervention (G×I) studies, which include random assignment to intervention condition, eliminate a common challenge with epidemiological G×E interaction studies, namely, that individuals do not usually randomly experience environments. Our genotypes influence the environments that we select into, often called gene–environment correlation, or rGE (Jaffee & Price, Reference Jaffee and Price2007; Keller, Reference Keller2014). Accordingly, G×I studies offer a unique opportunity to examine G×E because the environment is randomly assigned. This experimental element circumvents the problems that can confound G×E effects. In this way, G×I studies have the potential to uncover benefits of prevention and intervention for those at differing levels of genetic risk. Further, when differences in genetic vulnerability are not accounted for in intervention studies, analyses that collapse across all levels of genetic risk may mask differences in the effectiveness of interventions, and the results of prevention and intervention trials may be underestimated or misinterpreted. G×I studies provide a strong design to comprehensively understand both genetic and environmental influences on SUDs.
In this review, we summarize findings from G×I research on alcohol and other substance use behaviors, focusing on study design, sample composition, type of intervention, measurement of outcome, measurement of genotype, statistical methods, and main study findings. Review of this body of literature reveals themes that emerge across study findings for specific substances and G×I effects. We discuss the current state of the G×I literature with respect to measurement of genotype and outcome, study design, sample characteristics, and continued advances in the field of genetics. We conclude by providing recommendations for future research aimed at incorporating genetic data into intervention studies and argue for the importance of this type of research in the movement toward precision medicine.
Methods
Search procedure and review parameters
A systematic review of peer-reviewed literature in major scholarly databases, including PsycINFO, PubMed, and Google Scholar, was conducted. Searches included combinations of genetics terms (gene, genetics, polymorphism, single nucleotide polymorphism (SNP), genotype), substance use terms (alcohol, alcohol use, alcohol abuse, drinking, marijuana, cannabis, tobacco, smoking, nicotine, drug use, substance use), and intervention terms (intervention, prevention, treatment). Relevant Medical Subject Headings (MeSH) and Index terms were used in PubMed and PsycINFO, respectively, to ascertain studies within these broader literature categories. Reference sections of included articles as well as related meta-analytic reviews were screened to identify any additional studies for inclusion. The search yielded 1,195 unique articles for abstract screening. Studies included in this search were published in English in peer-review journals through April 2019. Included studies met the following inclusion criteria: (a) examined alcohol or other substance use as a quantitative outcome, (b) randomly assigned subjects to intervention or (active or inactive) control condition, (b) interventions were non-pharmacological psychosocial interventions for alcohol or other substance use, and (c) tested interactions between genotypes and intervention condition. Random assignment to the intervention and control conditions was required in order to control for the gene–environment correlation. Studies involving any pharmacological intervention, including nicotine replacement therapy, were excluded from the review as the goal of this systematic review was to examine interactions between genetic and environmental (rather than pharmacological) influences on patterns of behavior change.
An overview of the search and screening procedure is provided in Figure 1. Abstracts of all 1,195 studies identified in the search were screened for potential inclusion. A total of 44 studies advanced to full article screening, of which 27 were excluded. Reasons for exclusion were as follows: inclusion of a pharmacological intervention component (n = 9), did not examine alcohol or substance use as an outcome (n = 3), combined substance use with other risk behaviors (i.e., sexual behavior, n = 3), did not include random allocation to treatment (n = 4), did not examine G×I interaction effects (n = 3), did not include an intervention (n = 1), or the intervention involved genetic feedback (n = 4). Although interventions involving personalized genetic feedback may be beneficial in their own right, they address a different scientific question, namely how personalized feedback rather than underlying genetic risk influences outcomes. All studies were reviewed for extraction of the following variables: (a) author names and publication year, (b) study design, (c) sample characteristics, (d) intervention type, (e) outcome of interest, (f) genotype, and (g) G×I findings.
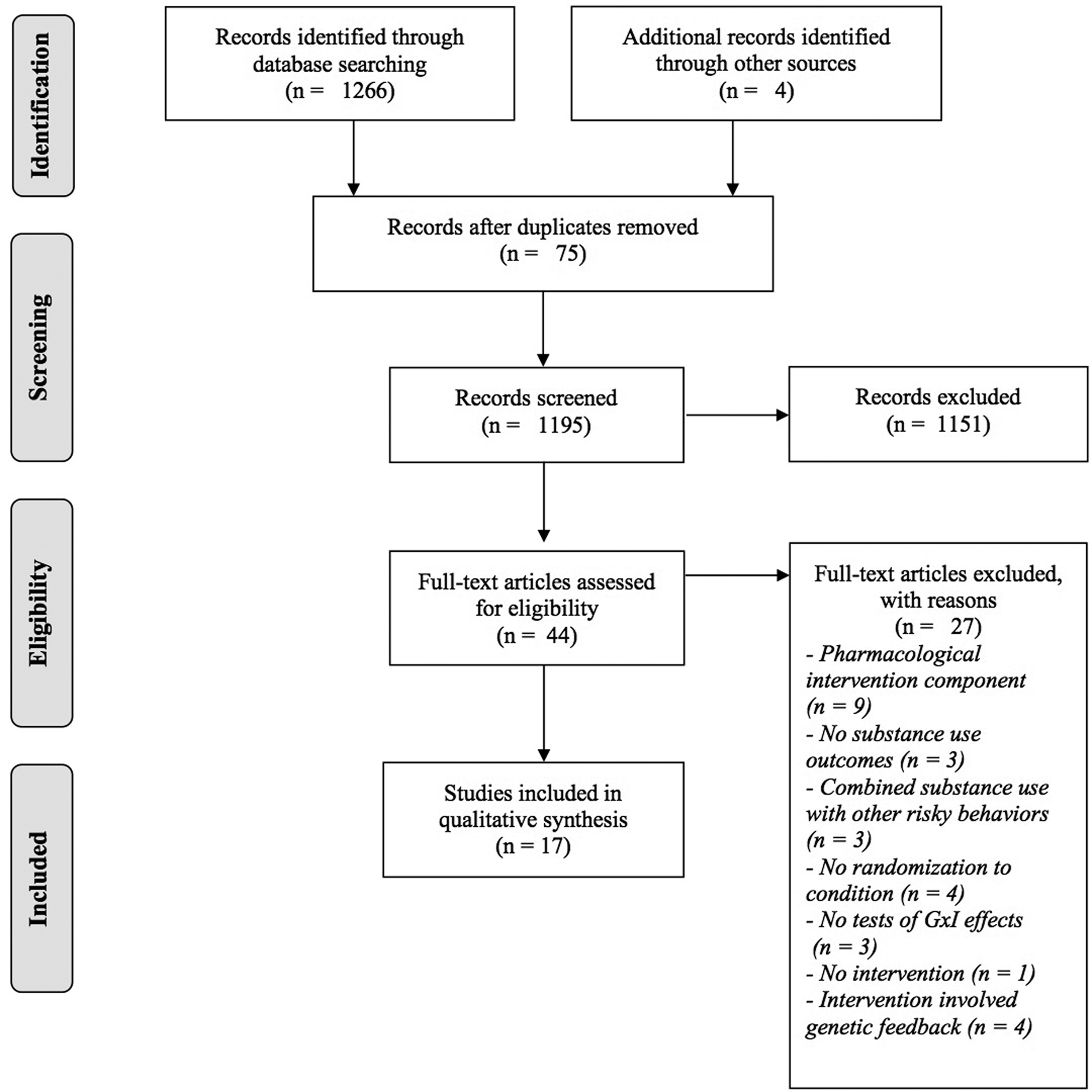
Figure 1. Overview of search and screening procedures
Results
Seventeen studies met criteria for inclusion in this review of G×I studies on alcohol and other substance use. Table 1 provides an overview of each study's design, sample characteristics, interventions, measured genotype, and outcome of interest. Below, the findings are summarized to elucidate patterns and themes across the included studies.
Table 1. Description of the 17 studies included in the review

Note: RCT = randomized controlled trial. ADH1B = alcohol dehydrogenase 1B, ADH1C = alcohol dehydrogenase 1C, ADH4 = alcohol dehydrogenase 4, ANKK1 = ankyrin repeat and kinase domain-containing 1, CHRNA3 = neuronal acetylcholine receptor subunit α-3, CHRNA5 = neuronal acetylcholine receptor subunit α-5, CHRNB4 = neuronal acetylcholine receptor subunit β−4, DRD2 = dopamine receptor D2, DRD4 = dopamine receptor D4, GABRA2 = GABA receptor subunit alpha-2, GABRG1 = GABA receptor subunit γ-1, 5-HTTLPR = serotonin transporter linked polymorphic region, MAOA = monoamine oxidase A, NR3C1 = nuclear receptor subfamily 3, group C, member 1, OXTR = oxytocin receptor
Study design and sample characteristics
Study design
There was considerable overlap in the research design and samples in the studies reviewed. Among the 17 studies, there were only nine unique projects represented. All of the projects used randomization to allocate participants to intervention or control conditions. Most of the projects (n = 6) involved youth recruited through large-scale prevention trials. Another project focused on prevention of alcohol problems in emerging adults, and the two remaining projects involved treatment-seeking populations in a clinical setting. Thus, with the exception of the two clinical samples described, the included studies focused largely on the prevention of risky behaviors rather than the treatment of SUDs. Prevention studies tended to have longer follow-up periods, ranging from 2–11 years for all but one study (Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009), which conducted a one-month follow-up. The clinical samples, in contrast, followed participants for no more than one year after the intervention.
Sample
Sample sizes ranged from 75 to 1,920 subjects, with variation across both prevention and clinical samples. Only two samples (Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009; Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016) included fewer than 100 subjects, with the remaining samples including 291 or more subjects. Studies involving the PROSPER sample (Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015; Cleveland, Griffin, et al., Reference Cleveland, Griffin, Wolf, Wiebe, Schlomer, Feinberg and Vandenbergh2018a; Cleveland, Schlomer, et al., Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b; Russell et al., Reference Russell, Schlomer, Cleveland, Feinberg, Greenberg, Spoth and Vandenbergh2018; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017) were the largest, exceeding 1,000 participants. Geographically, the studies included urban and rural communities across the US. Fast Track, Project MATCH, and PROSPER were multisite trials with samples from four, eight, and 28 communities, respectively. The Brody et al. (Reference Beach, Brody, Lei and Philibert2010, Reference Brody, Chen and Beach2013, Reference Brody, Yu and Beach2015) and Beach, Brody, Lei, and Philibert (Reference Beach, Brody, Lei and Philibert2010) samples were all recruited from a low-income community in rural Georgia. The remaining two studies were recruited from urban locations in Rhode Island (Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016) and Maryland (Musci et al., Reference Musci, Masyn, Uhl, Maher, Kellam and Ialongo2015, Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018).
Race/Ethnicity
Two studies included only European Americans (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015) and four studies included only African Americans recruited from rural Georgia (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007; Brody et al., Reference Brody, Chen and Beach2013, Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014, Reference Brody, Yu and Beach2015). Two studies had approximately equal groups of European American and African American participants (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018), and the remaining nine studies were predominantly European American (77–90%). One of these studies (Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009) reported that the sample was 87% Caucasian, but did not report the racial/ethnic background of the remaining 13% of the sample.
Sex
Studies also varied in the distribution of males and females in the samples. One study, which included interventions for intimate partner violence, was exclusively male (Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016). The remaining studies ranged from 40–73% male, with most studies including relatively balanced groups of male and female participants.
Age
Most of the studies included in this review recruited participants during adolescence (ages 11–17) and followed them longitudinally for several years. The Fast Track sample (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Zheng et al. Reference Zheng, Albert, McMahon, Dodge and Dick2018), and the Musci et al. (Reference Musci, Masyn, Uhl, Maher, Kellam and Ialongo2015, Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018) studies recruited students in early elementary school (kindergarten and first grade, respectively) and followed them longitudinally through young adulthood (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Musci et al., Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018). Ewing et al. (Reference Ewing, LaChance, Bryan and Hutchison2009) recruited emerging adults in a college setting. The remaining two studies included middle-aged adults (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007; Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016). See Table 1 for additional details on sample ages.
Interventions
School-based interventions
The Prevention Intervention Research Center at John Hopkins (Musci et al., Reference Musci, Masyn, Uhl, Maher, Kellam and Ialongo2015, Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018) implemented a school-based intervention during first grade that included curriculum changes, coaching teachers on instructional and behavior management practices, and focused strategies for underperforming children with the goal to prevent risk behaviors and reduce poor academic achievement, and disruptive/aggressive behavior in youth.
Family-centered interventions
Researchers at the Center for Family Research at the University of Georgia developed three family-centered interventions to prevent substance use and risky sexual behavior in youth. Strengthening African American Families (SAAF) recruited 11-year-olds and their parents to participate in seven weekly sessions to prevent the initiation of alcohol use and other risky behaviors (Beach et al., Reference Beach, Brody, Lei and Philibert2010; Brody et al., Reference Brody, Chen and Beach2013). The program included a shared meal, as well as separate and joint family groups focused on parenting behaviors, stress, peer pressure, and helping children appreciate their parents. SAAF was the foundation for the other two programs, Strengthening African American Families—Teen (SAAF-T) and Adults in the Making (AIM). SAAF-T was administered to teenagers (ages 14–16) and their parents for five weeks, with additional content on preventing risky sexual behavior (Brody et al., Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014). The AIM program targeted the prevention of risky substance use in 17-year-olds (Brody et al., Reference Brody, Yu and Beach2015). AIM comprised five weekly sessions for teens and parents, with the content focused on the transition from teenager to adult and strategies for racial socialization and discrimination.
Multicomponent interventions
Two projects administered multicomponent interventions with school-based and family-centered elements. First, Fast Track targeted students who were at risk for disruptive/aggressive behaviors to prevent conduct problems and externalizing behavior into adulthood (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018). They implemented a comprehensive program involving universal school curriculum, tutoring, home visits, group skills training, mentoring and individual services for children in the Fast Track project. The PROSPER project began with a universal family-focused intervention in 6th grade, followed by an in-school program in 7th grade (Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015; Cleveland et al., Reference Cleveland, Griffin, Wolf, Wiebe, Schlomer, Feinberg and Vandenbergh2018a; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017). The intervention communities chose among different options for both the family-focused and in-school components. All of the communities selected the “Strengthening Families Program” for the family-focused component, whereas three different programs were selected for the in-school component of the study and later controlled for in analyses.
Brief alcohol interventions
Two projects administered brief alcohol interventions (Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009; Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016). Brief alcohol interventions typically include assessment of substance use behaviors, personalized normative feedback on how one's own substance use compares to that of his/her peers, and motivational interviewing techniques to enhance readiness-to-change substance use behavior (Larimer & Cronce, Reference Larimer and Cronce2007).
Treatment of alcohol use disorders
One study included participants from Project MATCH, a multi-site, randomized controlled trial (RCT) of three psychosocial interventions for AUDs, with the original goal of identifying factors to match patients to the most effective intervention program (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007). The study included parallel arms of outpatient and after-care patients who were randomized to receive 12 weeks of individual, manualized motivational enhancement therapy (MET), cognitive-behavior therapy (CBT), or twelve-step facilitation (TSF).
Measurement of substance use
Outcome measures
Alcohol was the most common substance of focus, included in 14 of the 17 studies (see Table 1 for details). Six studies examined marijuana use, four studies examined cigarette smoking, and only one study assessed other substance use (e.g., Lysergic acid diethylamide (LSD), ecstasy, mushrooms, etc.) and nonmedical use of prescription drugs. Several studies combined across variables to create an index of substance use outcomes. For example, Brody et al. (Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014, Reference Brody, Yu and Beach2015) created a substance use index by aggregating frequency items for past month smoking, drinking, heavy drinking, and marijuana use. Overall, there was substantial variation in both the measurement of substance use and the handling of these variables in analyses (aggregate scores vs. individual measures) across the studies.
Data collection methods
Data were collected via structured and semi-structured interviews, paper and pencil self-report measures, computerized assessments, or a combination of these methods. Some projects used multiple reporters, including parents, teachers, and nominated peers. Overall, studies appeared to rely on standardized and validated measures with appropriate steps taken to assure participants of the confidentiality of their substance use data.
Measurement of genotype
Genetic variants
Most studies used candidate gene methods for measuring genetic factors. Candidate gene studies involve the study of a marker or markers located in a single gene selected a priori for its hypothesized role in a given phenotype, such as substance use behaviors (Dick et al., Reference Dick, Agrawal, Keller, Adkins, Aliev, Monroe and Sher2015). Variations in that marker, for example in the number of tandem repeats or alleles (e.g., AA, AG, or GG), are then tested for their association with an outcome of interest. A total of 45 different variants across 15 different genes were investigated in the reviewed G×I studies. The most commonly studied genetic variants were a variable number tandem repeat in dopamine receptor D4 (DRD4) and the serotonin transporter linked polymorphic region known as 5-HTTLPR. DRD4 is responsible for coding amino acids in the D4 subtype of the dopamine receptor (McGeary, Reference McGeary2009), and 5-HTTLPR plays a role in coding serotonin transporters (McHugh, Hofmann, Asnaani, Sawyer, & Otto, Reference McHugh, Hofmann, Asnaani, Sawyer and Otto2010). GABA receptor subunit alpha-2 (GABRA2), which was originally associated with alcohol dependence through linkage and association (Edenberg et al., Reference Edenberg, Dick, Xuei, Tian, Almasy, Bauer and Begleiter2004), was also a common target of research, appearing in three studies. One study that focused exclusively on smoking examined three nicotinic acetylcholine receptor subunit genes: neuronal acetylcholine receptor subunit α-5 (CHRNA5), neuronal acetylcholine receptor subunit α-3 (CHRNA3), and neuronal acetylcholine receptor subunit β-4 (CHRNB4) (Vandenbergh et al., Reference Vandenbergh, Schlomer, Cleveland, Schink, Hair, Feinberg and Redmond2015). Although most of the candidate gene studies examined single variants in separate models, some studies attempted to combine the effects of several single nucleotide polymorphisms (SNPs) into an aggregate measure of genotype. For example, one study averaged the effect of five SNPs in the OXTR gene, which codes for oxytocin receptors (Cleveland, Griffin, et al., Reference Cleveland, Griffin, Wolf, Wiebe, Schlomer, Feinberg and Vandenbergh2018a). Three other studies combined the effect of two to six variants from multiple genes into multi-locus (Brody et al., Reference Brody, Chen and Beach2013), multi-SNP (Cleveland, Schlomer, et al., Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b), or cumulative genetic risk scores (Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016).
Polygenic scores
Two studies used a polygenic scoring approach to measure genetic factors (Musci et al., Reference Musci, Masyn, Uhl, Maher, Kellam and Ialongo2015, Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018). Using results from genome-wide association studies, which systematically test for associations between millions of SNPs across the entire genome and an outcome of interest, genome-wide polygenic scores (GPS; also referred to as polygenic risk scores) can be created in independent samples by summing across the number of variants associated with the outcome weighted by the effect size drawn from the discovery genome-wide association studies (GWAS). The reviewed G×I studies that employed this approach derived polygenic scores for smoking quit success using a discovery sample GWAS of 550 European-Americans from three smoking cessation clinical trials (Uhl et al., Reference Uhl, Liu, Drgon, Johnson, Walther, Rose and Lerman2008). The Musci et al. (Reference Musci, Masyn, Uhl, Maher, Kellam and Ialongo2015, Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018) polygenic scores were created by summing across variants that conveyed greater success quitting cigarette smoking in the original trials (Uhl et al., Reference Uhl, Liu, Drgon, Johnson, Walther, Rose and Lerman2008). The derived quit success scores were associated with lower tobacco and marijuana use in youth and adults (Uhl et al., Reference Uhl, Walther, Musci, Fisher, Anthony, Storr and Rose2014). The discovery sample (N = 550) used to create the polygenic scores for these studies is now known to be highly underpowered. Current recommendations indicate that discovery samples in the hundreds of thousands are necessary to reliably identify genetic variants associated with complex behaviors (Dudbridge, Pashayan, & Yang, Reference Dudbridge, Pashayan and Yang2018).
Data collection methods and genotyping
DNA samples were collected via saliva for five studies (Beach et al., Reference Beach, Brody, Lei and Philibert2010; Brody et al., Reference Brody, Chen and Beach2013, Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014, Reference Brody, Yu and Beach2015; Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016), buccal cells for nine papers (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015; Cleveland et al., Reference Cleveland, Griffin, Wolf, Wiebe, Schlomer, Feinberg and Vandenbergh2018a; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b; Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009; Russell et al., Reference Russell, Schlomer, Cleveland, Feinberg, Greenberg, Spoth and Vandenbergh2018; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017; Vandenbergh et al., Reference Vandenbergh, Schlomer, Cleveland, Schink, Hair, Feinberg and Redmond2015; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018), and blood for one paper (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007). Two studies did not report how the DNA samples were collected (Musci et al., Reference Musci, Masyn, Uhl, Maher, Kellam and Ialongo2015, Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018). There was considerable variation in the amount of information provided about the genotyping procedures, but most studies reported use of either TaqMan to carry out quantitative polymerase chain reaction or Affymetrix arrays. Tests of Hardy–Weinberg equilibrium were also reported in most studies, with no evidence indicating that genotype or allele frequency in the samples differed from expected.
Statistical methods
Analytic design
Several different analytic approaches were used for tests of G×I including growth curve modeling (Brody et al., Reference Brody, Yu and Beach2015; Cleveland, Schlomer, et al., Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018), analysis of covariance (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017), and generalized linear models with log-linear models (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007), Poisson regression models (Brody et al., Reference Brody, Chen and Beach2013), and negative binomial regression (Brody et al., Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014) to account for unusual distributions in count data. Two studies used survival analysis (Musci et al., Reference Musci, Masyn, Uhl, Maher, Kellam and Ialongo2015, Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018), and one study used time-varying effect modeling (Russell et al., Reference Russell, Schlomer, Cleveland, Feinberg, Greenberg, Spoth and Vandenbergh2018). Several studies also employed multilevel modeling frameworks to account for nested data structures (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Beach et al., Reference Beach, Brody, Lei and Philibert2010; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015; Cleveland et al., Reference Cleveland, Griffin, Wolf, Wiebe, Schlomer, Feinberg and Vandenbergh2018a; Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016; Vandenbergh et al., Reference Vandenbergh, Schlomer, Cleveland, Schink, Hair, Feinberg and Redmond2015).
Race/ethnicity or genetic ancestry
Population stratification refers to systematic variations in allele frequency among different ancestral groups (Cardon & Palmer, Reference Cardon and Palmer2003). Many individuals also have mixed genetic ancestry (referred to as admixture), which can contribute to additional variation in allele frequency. When population stratification and admixture are not adjusted for in genetic analyses there is an increased risk of false-positive results (Hellwege et al., Reference Hellwege, Keaton, Giri, Gao, Velez Edwards and Edwards2017). Principal coordinates analysis (PCoA), principal components analysis (PCA), and multidimensional scaling (MDS) are all methods that use the distance between alleles to map the structure of different populations (Halder, Shriver, Thomas, Fernandez, & Frudakis, Reference Halder, Shriver, Thomas, Fernandez and Frudakis2008; Price et al., Reference Price, Patterson, Plenge, Weinblatt, Shadick and Reich2006). These methods provide a series of principal components (PCs), which contain the primary factors that differentiate ancestral groups. The inclusion of genetic ancestry PCs in genetic analyses adjusts for population stratification as well as admixture.
Across the studies reviewed, eight papers included PCs derived from PCoA, PCA, or MDS in their analyses to minimize population stratification (Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015; Cleveland et al., Reference Cleveland, Griffin, Wolf, Wiebe, Schlomer, Feinberg and Vandenbergh2018a; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b; Musci et al., Reference Musci, Masyn, Uhl, Maher, Kellam and Ialongo2015, Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018; Russell et al., Reference Russell, Schlomer, Cleveland, Feinberg, Greenberg, Spoth and Vandenbergh2018; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017; Vandenbergh et al., Reference Vandenbergh, Schlomer, Cleveland, Schink, Hair, Feinberg and Redmond2015). Two studies conducted their analyses separately for European Americans and African Americans based on self-reported race/ethnicity (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018). One study covaried for self-report race/ethnicity with a dummy-coded variable representing Caucasian (86%) versus non-Caucasian (14%) (Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009). Stuart et al. (Reference Stuart, McGeary, Shorey and Knopik2016) also reported including race as a covariate, but did not provide information as to how it was coded for analyses. Lastly, four studies included only participants who self-identified as African American, and did not control for population stratification or admixture in their analyses (Beach et al., Reference Beach, Brody, Lei and Philibert2010; Brody et al., Reference Brody, Chen and Beach2013, Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014, Reference Brody, Yu and Beach2015).
Tests of artifact interactions
None of the studies tested for artifact interactions, in which a genotype unrelated to the outcome of interest is included into the model in place of the target genotype (Dempfle et al., Reference Dempfle, Scherag, Hein, Beckmann, Chang-Claude and Schäfer2008). Tests of artifact interaction strengthen the validity of the study findings by confirming that the results do not replicate with a theoretically unrelated genotype.
Correction for multiple testing
Two studies reported use of corrections for multiple testing in G×I models that examined 10 SNPs on nuclear receptor subfamily 3, group C, member 1 (NR3C1) in two different racial-ethnic groups (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018). Another study, which examined 14 different variants on five different genes, used the false discovery rate to reduce the likelihood of incurring a Type I error (Brody et al., Reference Brody, Chen and Beach2013). No other studies reported use of methods to correct for multiple testing.
Main study findings
Main effect of intervention
Eight (47%) of the 17 reviewed studies found significant main effects of intervention indicating lower alcohol or other substance use outcomes for those in the intervention condition (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Beach et al., Reference Beach, Brody, Lei and Philibert2010; Brody et al., Reference Brody, Chen and Beach2013; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b; Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009; Musci et al., Reference Musci, Masyn, Uhl, Maher, Kellam and Ialongo2015; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017; Vandenbergh et al., Reference Vandenbergh, Schlomer, Cleveland, Schink, Hair, Feinberg and Redmond2015). Seven studies (41%) found no evidence for a significant effect of intervention (Brody et al., Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014, Reference Brody, Yu and Beach2015; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015; Cleveland et al., Reference Cleveland, Griffin, Wolf, Wiebe, Schlomer, Feinberg and Vandenbergh2018a; Musci et al., Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018; Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018). Two studies did not provide enough information to determine whether a significant main effect of intervention was present (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007; Russell et al., Reference Russell, Schlomer, Cleveland, Feinberg, Greenberg, Spoth and Vandenbergh2018).
Main effect of genotype
Seven (41%) of the studies found significant main effects of genetic factors on alcohol or other substance use outcomes (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007; Brody et al., Reference Brody, Chen and Beach2013, Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b; Musci et al., Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018; Vandenbergh et al., Reference Vandenbergh, Schlomer, Cleveland, Schink, Hair, Feinberg and Redmond2015). Specifically, SNP rs279858 of GABRA2 was associated with alcohol use (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007; Brody et al., Reference Brody, Chen and Beach2013) and heavy drinking (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007). The polymorphic region 5-HTTLPR as well as multi-SNP scores for alcohol dehydrogenase 1B (ADH1B) (rs1042026, rs1229984) and alcohol dehydrogenase 1B (ADH1C) (rs698, rs1614972) were also associated with alcohol use (Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b). Brody et al. (Reference Brody, Chen and Beach2013) observed significant main effects of genotype on alcohol use for all ten independent dopaminergic (DRD2, DRD4, ankyrin repeat and kinase domain containing 1 (ANKK1)) and GABAergic (GABA receptor subunit γ-1, GABRA2) variants examined. Musci et al. observed a significant main effect of polygenic score for quit success on age of first marijuana use (2018) but not age of first tobacco use (2015). Seven other studies (41%) found no evidence for significant main effects of genotype. Studies with null findings examined variants in DRD4 (Beach et al., Reference Beach, Brody, Lei and Philibert2010; Brody et al., Reference Brody, Yu and Beach2015), 5-HTTLPR (Beach et al., Reference Beach, Brody, Lei and Philibert2010; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017; Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016), OXTR (Cleveland, Griffin, et al., Reference Cleveland, Griffin, Wolf, Wiebe, Schlomer, Feinberg and Vandenbergh2018a), and MAOA (Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016). Three studies did not provide enough information to determine whether significant main effects of genotype were present (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009; Russell et al., Reference Russell, Schlomer, Cleveland, Feinberg, Greenberg, Spoth and Vandenbergh2018).
G×I interaction effects
All studies reported at least one significant G×I effect in the form of either two-way (15 studies) or three-way interactions (two studies), detailed in Table 2.
Table 2. Results of gene-by-intervention (G×I) analyses

Note: ADH1C = alcohol dehydrogenase 1C, DRD4 = dopamine receptor D4, GABRA2 = GABA receptor subunit alpha-2, 5-HTTLPR = serotonin transporter linked polymorphic region, MAOA = monoamine oxidase A, NR3C1 = nuclear receptor subfamily 3, group C, member 1, OXTR = oxytocin receptor
Several studies observed significant interactions between intervention condition and SNPs within the dopaminergic system, particularly the variable number tandem repeat (VNTR) in DRD4. Three studies found that individuals with the long DRD4 allele (7+ repeats) responded better to intervention as indicated by lower substance use index scores (Beach et al., Reference Beach, Brody, Lei and Philibert2010; Brody et al., Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014, Reference Brody, Yu and Beach2015). In contrast, Ewing et al. (Reference Ewing, LaChance, Bryan and Hutchison2009) observed that DRD4 short allele carriers repeat carriers took more steps to change their drinking. Cleveland et al. (Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015) found no evidence for a two-way interaction between DRD4 and intervention condition; however, they observed a three-way interactions between DRD4, intervention condition, and maternal involvement such that carriers of the DRD4 7-repeat allele with high maternal involvement in the intervention condition showed significantly less initiation of alcohol use and drunkenness than their counterparts in the control group. Brody et al. (Reference Brody, Chen and Beach2013) found a significant interaction between intervention condition and a SNP in DRD2 influencing alcohol use, but found no interaction effects for DRD4 or ANKK1 variants, the latter of which is involved in dopamine receptor density.
Three studies detected interactions between intervention condition and SNPs within GABAergic genes (gamma-aminobutyric acid type A receptor subunit gamma1 [GABRG1], GABRA2) influencing adolescent alcohol use (Brody et al., Reference Brody, Chen and Beach2013), adolescent alcohol misuse (Russell et al., Reference Russell, Schlomer, Cleveland, Feinberg, Greenberg, Spoth and Vandenbergh2018), and adult daily drinking and any drinking (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007). Three studies of the serotonergic gene system (5-HTTLPR, monoamine oxidase A (MAOA)) detected significant G×I effects influencing alcohol initiation (Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015), adolescent substance misuse initiation (Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017) and percent days abstinent in an adult clinical sample (Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016). Two studies found that SNPs on the glucocorticoid receptor gene nuclear receptor subfamily 3, group C, member 1 (NR3C1) interacted with intervention condition to influence problem alcohol and cannabis use (rs10482672; Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015), and alcohol abuse (rs12655166; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018).
Cleveland, Griffin et al. (Reference Cleveland, Griffin, Wolf, Wiebe, Schlomer, Feinberg and Vandenbergh2018a) examined five SNPs on the oxytocin receptor gene OXTR and found a significant three-way interaction between OXTR multi-SNP scores, intervention condition, and peer substance use on rates of alcohol use. Cleveland, Schlomer et al. (Reference Cleveland, Schlomer, Vandenbergh, Wolf, Feinberg, Greenberg and Redmond2018b) studied six SNPs located on three alcohol dehydrogenase encoding genes (ADH1B, ADH1C, alcohol dehydrogenase 4 (ADH4)) and found significant G×I effects for ADH1C on alcohol use. Vandenbergh et al. (Reference Vandenbergh, Schlomer, Cleveland, Schink, Hair, Feinberg and Redmond2015) found that rs16969968 in CHRNA5 interacted with intervention condition to influence past month cigarette smoking. Finally, Musci et al. (Reference Musci, Fairman, Masyn, Uhl, Maher, Sisto and Ialongo2018) observed significant G×I effects on age of first marijuana use and tobacco use (2015) using a polygenic score for quit success, with higher polygenic score predicting later initiation. As noted previously, a total of 45 different genetic variants were examined in the reviewed studies. Thus, the significant G×I effects described above were accompanied by reports of null G×I findings in most (76%) papers as well. Null findings occurred both with other genetic variants on the same outcomes, as well as the same variants across multiple outcomes. Only four studies (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007; Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009; Russell et al., Reference Russell, Schlomer, Cleveland, Feinberg, Greenberg, Spoth and Vandenbergh2018; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017) did not report null G×I findings. Despite the relative prevalence of null findings in the reviewed studies, there is some indication that publication bias may exist given that all papers reported at least one significant G×I finding. Further, only one study (Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017) was designed as an explicit attempt to replicate previous G×I findings. Schlomer et al. (Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017) successfully replicated the G×I finding, but failed to replicate the main effect of 5-HTTLPR observed in the original study. Other studies were successful in generalizing (rather than replicating) the DRD4 findings in different populations (Brody et al., Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014, Reference Brody, Yu and Beach2015; Cleveland et al., Reference Cleveland, Schlomer, Vandenbergh, Feinberg, Greenberg, Spoth and Hair2015). All other studies represent novel tests of G×I interaction effects on alcohol and other substance use.
Discussion
The purpose of this systematic review was to characterize the current state of the literature on G×I studies of alcohol and other substance use behaviors, with particular attention to trends in results and methodological approaches used across the examined studies. The conclusions drawn from this review are based on the 17 studies that met criteria for inclusion. A number of themes emerged from the review of the extant literature. Below, we discuss each of them in turn, and provide recommendations for advancing the field of G×I research moving forward.
The choice of “G” in G×I studies
The most notable finding from this review was that almost all G×I studies of alcohol and other substance use outcomes examined candidate genes. Many of the early G×I studies examined candidates selected based on hypothesized biological rationale, for example DRD2, DRD4, MAOA, and 5-HTTLPR (Beach et al., Reference Beach, Brody, Lei and Philibert2010; Brody et al., Reference Brody, Chen and Beach2013; Ewing et al., Reference Ewing, LaChance, Bryan and Hutchison2009; Stuart et al., Reference Stuart, McGeary, Shorey and Knopik2016). Other studies selected candidates derived from early linkage studies such as GABRA2 (Bauer et al., Reference Bauer, Covault, Harel, Das, Gelernter, Anton and Kranzler2007; Brody et al., Reference Brody, Chen and Beach2013; Edenberg et al., Reference Edenberg, Dick, Xuei, Tian, Almasy, Bauer and Begleiter2004). In this way, the history of incorporating genetic information into prevention science has paralleled the history of genetics, which originally focused on more targeted genotyping (candidate genes), subsequently moved to genome-wide scans with sparse coverage (linkage studies), and more recently has moved to genome-wide association studies (GWAS), which systematically and atheoretically test for associations between millions of SNPs across the entire genome and an outcome of interest (Duncan & Keller, Reference Duncan and Keller2011; Purcell et al., Reference Purcell, Wray, Stone, Visscher, O'Donovan, Sullivan and Sklar2009; Wray et al., Reference Wray, Lee, Mehta, Vinkhuyzen, Dudbridge and Middeldorp2014). To accommodate very small effect sizes and multiple testing, very large samples and stringent p value thresholds are necessary to obtain adequate statistical power in GWAS (Hong & Park, Reference Hong and Park2012).
Extensive efforts are now underway to amass very large samples in order to identify the specific genetic variants that contribute to risk for substance use and SUDs (Cabana-Domínguez, Shivalikanjli, Fernàndez-Castillo, & Cormand, Reference Cabana-Domínguez, Shivalikanjli, Fernàndez-Castillo and Cormand2019; Cheng et al., Reference Cheng, Zhou, Sherva, Farrer, Kranzler and Gelernter2018; Liu et al., Reference Liu, Jiang, Wedow, Li, Brazel, Chen and Vrieze2019; Pasman et al., Reference Pasman, Verweij, Gerring, Stringer, Sanchez-Roige, Treur and Vink2018). The most recent GWAS of substance use and dependence include samples ranging from the hundreds of thousands to 1.2 million individuals (Clarke et al., Reference Clarke, Adams, Davies, Howard, Hall, Padmanabhan and McIntosh2017; Kranzler et al., Reference Kranzler, Zhou, Kember, Smith, Justice, Damrauer and Gelernter2019; Liu et al., Reference Liu, Jiang, Wedow, Li, Brazel, Chen and Vrieze2019; Polimanti et al., Reference Polimanti, Peterson, Ong, MacGregor, Edwards, Clarke and Derks2019; Sanchez-Roige et al., Reference Sanchez-Roige, Palmer, Fontanillas, Elson, Adams and Clarke2019), the largest of which (Liu et al., Reference Liu, Jiang, Wedow, Li, Brazel, Chen and Vrieze2019) identified 566 genetic variants associated with alcohol consumption and smoking. Results of these well-powered GWAS examining hundreds of thousands or millions of individuals largely have not supported the hypothesized role of suspected candidate genes across a variety of psychiatric outcomes (Border et al., Reference Border, Johnson, Evans, Smolen, Berley, Sullivan and Keller2019; Johnson et al., Reference Johnson, Border, Melroy-Greif, de Leeuw, Ehringer and Keller2017; Van der Auwera et al., Reference Van der Auwera, Peyrot, Milaneschi, Hertel, Baune, Breen and Grabe2018). Systematic reviews of the candidate gene literature also found that candidate gene studies were underpowered, susceptible to publication bias, and rarely replicated across studies (Border et al., Reference Border, Johnson, Evans, Smolen, Berley, Sullivan and Keller2019; Dick et al., Reference Dick, Agrawal, Keller, Adkins, Aliev, Monroe and Sher2015; Duncan & Keller, Reference Duncan and Keller2011). For these reasons, the early candidate gene studies are now considered problematic (Latendresse, Musci, & Maher, Reference Latendresse, Musci and Maher2018; Musci & Schlomer, Reference Musci and Schlomer2018). One exception is the alcohol dehydrogenase (ADH) genes which emerged as biological candidates for alcohol use outcomes based on their known role in ethanol metabolism (Edenberg, Reference Edenberg2007) and continue to be highly associated with alcohol dependence in genome-wide association studies (Clarke et al., Reference Clarke, Adams, Davies, Howard, Hall, Padmanabhan and McIntosh2017; Tawa et al., Reference Tawa, Hall and Lohoff2016; Walters et al., Reference Walters, Adams, Adkins, Aliev, Bacanu, Batzler and Chen2018). Aside from the ADH genes, the early enthusiasm for using hypothesis-driven candidate genes to explain the underlying etiology of complex behaviors has been diminished by the subsequent recognition that many significant candidate gene findings were likely false positives (Dick et al., Reference Dick, Barr, Cho, Cooke, Kuo, Lewis and Su2018). Simulations suggest that more than 70% of the genetic influence on complex behaviors can be attributed to the combined effect of many common loci of small effect (Moser et al., Reference Moser, Lee, Hayes, Goddard, Wray and Visscher2015). By testing for the effect of a single variant, candidate gene studies were inconsistent with the evidence that genetic influences are best understood as an accumulation of multiple small genetic effects, with very little variance explained by any single variant (Timpson, Greenwood, Soranzo, Lawson, & Richards, Reference Timpson, Greenwood, Soranzo, Lawson and Richards2018).
With these new insights, the field of genetics moved towards methods that capitalize on findings from GWAS to index broad genetic vulnerability across the genome (Bogdan, Baranger, & Agrawal, Reference Bogdan, Baranger and Agrawal2018; Duncan & Keller, Reference Duncan and Keller2011; Purcell et al., Reference Purcell, Wray, Stone, Visscher, O'Donovan, Sullivan and Sklar2009; Wray et al., Reference Wray, Lee, Mehta, Vinkhuyzen, Dudbridge and Middeldorp2014). By creating genome-wide polygenic scores (also referred to as polygenic risk scores), we can now capture risk across the multitude of genetic variants involved in complex behaviors using a single continuous aggregate score (Purcell et al., Reference Purcell, Wray, Stone, Visscher, O'Donovan, Sullivan and Sklar2009; Wray, Goddard, & Visscher, Reference Wray, Goddard and Visscher2007). There are multiple techniques for calculating genome-wide polygenic scores (GPS), but the method broadly involves summing the number of alleles for each SNP meeting a specified significance threshold, weighted according to their effect size determined by their association with a given outcome in a well-powered, independent GWAS sample. Higher scores indicate a greater genetic predisposition for the outcome of interest. A number of existing papers provide thorough review of the conceptual meaning, calculation methods, and clinical utility of polygenic scores (Bogdan et al., Reference Bogdan, Baranger and Agrawal2018; Dudbridge, Reference Dudbridge2016; Maier, Visscher, Robinson, & Wray, Reference Maier, Visscher, Robinson and Wray2018; Wray et al., Reference Wray, Lee, Mehta, Vinkhuyzen, Dudbridge and Middeldorp2014). Across studies of alcohol and other substance use, GPS produce consistent, yet modest effect sizes (Chang et al., Reference Chang, Couvy-Duchesne, Liu, Medland, Verhulst and Benotsch2019; Kranzler et al., Reference Kranzler, Zhou, Kember, Smith, Justice, Damrauer and Gelernter2019; Linnér et al., Reference Linnér, Biroli, Kong, Meddens, Wedow, Fontana and Beauchamp2019; Liu et al., Reference Liu, Jiang, Wedow, Li, Brazel, Chen and Vrieze2019; Pasman et al., Reference Pasman, Verweij, Gerring, Stringer, Sanchez-Roige, Treur and Vink2018; Walters et al., Reference Walters, Adams, Adkins, Aliev, Bacanu, Batzler and Chen2018), which are expected to improve as sample sizes for discovery GWAS increase (Maher, Reference Maher2015).
As indices of aggregate genetic vulnerability, GPS offer new opportunities for developmental research and prevention science. GPS can be calculated for any measurable construct believed to be associated with an outcome of interest for which a large discovery GWAS exists. Accordingly, developmental theories surrounding the processes that underlie the initiation and progression of substance use behaviors can be incorporated into research studies using GPS (Masten, Faden, Zucker, & Spear, Reference Masten, Faden, Zucker and Spear2009; Rogosch, Oshri, & Cicchetti, Reference Rogosch, Oshri and Cicchetti2010; Tarter, Reference Tarter2002). For example, by calculating a GPS for externalizing behaviors, a well-established pathway of risk to substance use problems (Colder et al., Reference Colder, Scalco, Trucco, Read, Lengua, Wieczorek and Hawk2013; King, Iacono, & McGue, Reference King, Iacono and McGue2004; Krueger et al., Reference Krueger, Hicks, Patrick, Carlson, Iacono and McGue2002), one could examine the influence of genetic risk toward externalizing behaviors on the unfolding of substance use behaviors across time. Using a G×I framework, researchers could then also capitalize on randomized prevention designs to evaluate the degree to which a youth prevention program targeting related behaviors (e.g., behavioral disinhibition, antisocial behaviors) might mitigate high genetic risk for externalizing problems. Initial efforts to use large-scale GWAS findings to develop polygenic scores and integrate them into G×I analyses in prevention samples are already underway. In a recently published paper, Kuo et al. (Reference Kuo, Salvatore, Aliev, Ha, Dishion and Dick2019) examined alcohol use outcomes in a G×I framework using polygenic scores derived from a published GWAS study of alcohol dependence of 5,131 European American and 4,629 African American subjects (Gelernter et al., Reference Gelernter, Kranzler, Sherva, Almasy, Koesterer, Smith and Anton2014). Findings indicated that a preventive intervention moderated the effect of polygenic risk associated with alcohol dependence, such that higher polygenic scores were associated with increased risk of alcohol dependence diagnosis in the control condition but not in the intervention condition among European American individuals. Outside of the intervention literature, other studies have also demonstrated significant G×E effects using polygenic scores (Barr et al., Reference Barr, Kuo, Aliev, Latvala, Viken, Rose and Dick2019; Pasman et al., Reference Pasman, Verweij, Abdellaoui, Hottenga, Fedko, Willemsen and Vink2020; Salvatore et al., Reference Salvatore, Aliev, Edwards, Evans, Macleod, Hickman and Dick2014). This research is consistent with patterns of results from the broader twin study literature suggesting that measures of aggregate genetic risk are useful for understanding G×E effects (Young-Wolff, Enoch, & Prescott, Reference Young-Wolff, Enoch and Prescott2011). Some environments are expected to exacerbate genetic risk, whereas others may mitigate genetic risk.
The use of polygenic scores is not without limitations, and may also pose challenges for developmental psychology and prevention science research. First, genome-wide data, which are required to calculated GPS, are more expensive to generate than the small number of markers typically genotyped in candidate genes. While the cost of genotyping continues to decrease, large-scale clinical trials may continue to find genotyping costs prohibitive for some time. Second, the predictive validity of polygenic scores remains relatively low, accounting for less than 5% of variance in substance use behaviors in the most current large, well-powered GWAS (Kranzler et al., Reference Kranzler, Zhou, Kember, Smith, Justice, Damrauer and Gelernter2019; Linnér et al., Reference Linnér, Biroli, Kong, Meddens, Wedow, Fontana and Beauchamp2019; Liu et al., Reference Liu, Jiang, Wedow, Li, Brazel, Chen and Vrieze2019; Pasman et al., Reference Pasman, Verweij, Gerring, Stringer, Sanchez-Roige, Treur and Vink2018). The amount of variance accounted for has been shown to increase dramatically with increases in discovery GWAS sample sizes (Dudbridge, Reference Dudbridge2013; Evangelou et al., Reference Evangelou, Warren, Mosen-Ansorena, Mifsud, Pazoki, Gao and Caulfield2018; Lee et al., Reference Lee, Wedow, Okbay, Kong, Maghzian, Zacher and Cesarini2018; Pardiñas et al., Reference Pardiñas, Holmans, Pocklington, Escott-Price, Ripke, Carrera and Walters2018; Wray et al., Reference Wray, Lee, Mehta, Vinkhuyzen, Dudbridge and Middeldorp2014); however, substance use research is not yet at that point. The predictive power of polygenic scores is directly related to the size and power of the GWAS sample used to create them. Thus, developmental scientists are limited to the calculation of GPS for phenotypes for which large, well-powered GWAS exist. Third, if individual variants within a polygenic score are moderated by the environment in different directions, the method of creating aggregate GPS based on GWAS results, and then testing for moderation of the environment, may miss important interactions. Methods are under development to test for SNP-level G×E effects across the genome (Coleman et al., Reference Coleman, Peyrot, Purves, Davis, Rayner, Choi and Breen2020; Polimanti et al., Reference Polimanti, Kaufman, Zhao, Kranzler, Ursano, Kessler and Stein2018). However, these analyses require very large sample sizes for adequate power (hundreds of thousands to millions of individuals), parallel to GWAS, with genotypic, phenotypic, and environmental data available, making this an unlikely option for intervention trials in the near future. Finally, genetic technology advances very rapidly, with new methods for calculating GPS emerging each year. Reviews about integrating genetics into prevention science (Belsky & Israel, Reference Belsky and Israel2014; Dick, Latendresse, & Riley, Reference Dick, Latendresse and Riley2011; Latendresse et al., Reference Latendresse, Musci and Maher2018) and communicating with genetics researchers may prove helpful, but the rapid developments in genetic technology remain challenging to accommodate or anticipate for those within and outside of the field of genetics. This suggests that partnerships between prevention researchers and geneticists will remain critical for prevention research to reflect the state of the science in genetics.
Mediators: understanding why G×I occurs
As trends in genetic technology move towards measures of aggregate genetic vulnerability, testing for theory-driven mediators and processes underlying G×I effects becomes increasingly important. Polygenic scores reflect a sum of genetic factors that predispose an individual toward substance use problems, and may include genetic variants that influence the outcome through a variety of pathways, such as sensation seeking, sociability, internalizing, etc. Therefore, mediation analyses can help uncover the mechanisms driving the relationship between polygenic scores and prevention/intervention outcomes. Two of the reviewed studies examined mediators of G×I effects (Brody et al., Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014, Reference Brody, Yu and Beach2015). The first study tested mediated moderation, examining changes in parenting practices as a function of the family-based SAAF-T program (Brody et al., Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014). The second study tested “vulnerability cognitions,” defined as thoughts that may increase the likelihood of substance use, such as “intentions to use drugs, willingness to use, and positive prototypes or images of drug-using peers” as a mechanism of the G×I effect (Brody et al., Reference Brody, Yu and Beach2015). Both studies found support for the mediation of G×I effects via improved parenting practices (Brody et al., Reference Brody, Chen, Beach, Kogan, Yu, Diclemente and Philibert2014) and reduced vulnerability cognitions (Brody et al., Reference Brody, Yu and Beach2015). These analyses took an important step toward answering the question of how the intervention differentially affects individuals with different genotypes. With rich training in theoretical models and mechanisms of behavior change, developmental researchers and clinical scientists are well-positioned to lead the field in identification and evaluation of concepts that are theoretically implicated in genetics, development, and prevention/intervention. For example, selection of peers is related to substance initiation, influenced by genetics (Kendler & Baker, Reference Kendler and Baker2007; Kendler et al., Reference Kendler, Jacobson, Gardner, Gillespie, Aggen and Prescott2007; Tarantino et al., Reference Tarantino, Tully, Garcia, South, Iacono and McGue2014), and can also be targeted in prevention programming (Dodge, Dishion, & Lansford, Reference Dodge, Dishion and Lansford2006; Hansen & Graham, Reference Hansen and Graham1991; Larimer & Cronce, Reference Larimer and Cronce2007). Studying theory-driven mediators of G×I effects may enable researchers to further identify factors that could be harnessed to enhance intervention effects.
Developmental considerations for G×I
Most of the reviewed G×I studies were conducted using prevention trials in youth, with only two studies testing G×I effects in adult clinical samples. Only one reviewed study focused on college-aged students, which is a known period of elevated risk for the development of substance use problems (Skidmore, Kaufman, & Crowell, Reference Skidmore, Kaufman and Crowell2016; Sussman & Arnett, Reference Sussman and Arnett2014). Additional research is needed on a range of developmental periods (including emerging, middle, and older adulthood) as well as additional focus on SUD treatment. The length of follow-up for adult samples tended to be shorter than the youth prevention samples. It would be beneficial to expand the range of follow-up beyond one year post-intervention, especially in light of high rates of recurrence among individuals with SUDs. In addition, some studies followed youth through age 25 (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Zheng et al., Reference Zheng, Albert, McMahon, Dodge and Dick2018), but there were no studies that spanned the period of youth through middle adulthood. The relative influence of genetic risk for alcohol and substance use varies across the lifespan, with robust evidence that genetic factors affecting substance dependence are less important in early adolescence, when environmental factors are predominant, and become more influential across adolescence into adulthood (Dick et al., Reference Dick, Pagan, Viken, Purcell, Kaprio, Pulkkinen and Rose2007, Reference Dick, Cho, Latendresse, Aliev, Nurnberger, Edenberg and Kuperman2014; Edwards & Kendler, Reference Edwards and Kendler2013; Kendler, Jacobson, Myers, & Eaves, Reference Kendler, Jacobson, Myers and Eaves2008; Meyers et al., Reference Meyers, Salvatore, Vuoksimaa, Korhonen, Pulkkinen, Rose and Dick2014). The impact of developmental changes in the context of intervention designs can be accommodated only with longitudinal designs that span these periods. Extending the longitudinal study of youth to include developmental periods into adulthood will allow us to understand the influence of genetics across the development. Growing G×I research with clinical adult samples will allow us to understand both short-term and long-term G×I effects on treatment response.
The outcomes we study
The reviewed research focused primarily on alcohol (14 reviewed studies), but other substance use was not extensively addressed. Marijuana was included in 35% of studies, smoking in 23% of studies, and only one study measured other substances (e.g., cocaine, non-medical use of prescription drugs). The expanding legalization of marijuana and the opioid overdose epidemic underscore the importance of understanding the role of genetics in prevention and treatment of these substances. Furthermore, the shared genetic risk for substance use and related behaviors indicates that G×I studies that assess a range of substances may better align with etiological theories (Glantz & Leshner, Reference Glantz and Leshner2000; Kendler, Prescott, Myers, & Neale, Reference Kendler, Prescott, Myers and Neale2003; Mayes & Suchman, Reference Mayes and Suchman2015; Vanyukov et al., Reference Vanyukov, Tarter, Kirisci, Kirillova, Maher and Clark2003, Reference Vanyukov, Tarter, Kirillova, Kirisci, Reynolds, Kreek and Ridenour2012). Future research that focuses on G×I effects in the prevention/intervention of marijuana and other illicit substances, in addition to alcohol, may more comprehensively address the problem of substance use and related harms.
In the reviewed studies, we also observed substantial variation in the measurement and construction of outcome variables across different domains of substance use/misuse (e.g., initiation, frequency of use, substance-related problems). Both shared and unique genetic factors are involved in risk for alcohol and substance-related behaviors, such as frequency of use, quantity of use, and age of initiation (Kendler et al., Reference Kendler, Prescott, Myers and Neale2003). In addition, alcohol consumption and alcohol problems only show partially overlapping genetic effects in GWAS (Kranzler et al., Reference Kranzler, Zhou, Kember, Smith, Justice, Damrauer and Gelernter2019; Walters et al., Reference Walters, Adams, Adkins, Aliev, Bacanu, Batzler and Chen2018). While composite measures of substance use behaviors may increase power and be theoretically justified for examining particular research questions, it is difficult to discern the specific impact of prevention/intervention effects with combined measures and thus examine mechanisms of G×I effects. Reporting findings for both composite measures and individual domains of substance use behaviors using standardized measures would enhance our ability to identify themes and compare findings across studies. A number of resources are available for selecting phenotypic measures that can be standardized across studies, such as the PhenX Toolkit (https://www.phenxtoolkit.org/), and the database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap).
Diversity in samples
The equitable application of G×I research findings is contingent upon the recruitment of samples that fully represent the diversity present in the US and around the world. Most of the reviewed study samples (N = 9) comprised predominantly European Americans, which is not representative of the diverse racial/ethnic composition of the broader US population. Although several studies included substantial proportions of African Americans, other minorities (e.g., Asian, Hispanic) were not well-represented in the G×I studies. It is also important to note that at present, there is considerable disparity between predictive power and utility of genome-wide polygenic scores for individuals of European ancestry compared to all other ancestry groups (Martin et al., Reference Martin, Kanai, Kamatani, Okada, Neale and Daly2019). A number of factors contribute to the underrepresentation of minority populations in genetic research (Dick, Barr, Guy, Nasim, & Scott, Reference Dick, Barr, Guy, Nasim and Scott2017), including smaller numbers from which to draw samples, computational complications of diverse samples in genomic research (Peterson et al., Reference Peterson, Edwards, Bacanu, Dick, Kendler and Webb2017), and the fact that many existing genetically-informative samples were recruited at a time when there was less attention to inclusion and diversity in research and more focus on creating homogenous groups (Haga, Reference Haga2010). There are also a number of sociocultural considerations that contribute to observed disparities, such as the history of the eugenics movement in the US, stigma, mistrust of science, and concerns about discrimination, abuse, and confidentiality (Bates, Lynch, Bevan, & Condit, Reference Bates, Lynch, Bevan and Condit2005; Furr, Reference Furr2002; Schulz, Caldwell, & Foster, Reference Schulz, Caldwell and Foster2003). The underrepresentation of individuals of non-European ancestry inhibits the utility and applicability of findings from G×I research, particularly in regards to the future of precision medicine (Scott, Reference Scott2017). Decreasing cost of genotyping and collaborative consortia (e.g., Psychiatric Genomics Consortium) and initiatives, such as the All of Us Research Program, which endeavors to collect data from a diverse sample of one million Americans, will help facilitate the collection of large, diverse samples.
One barrier to diversifying the racial/ethnic composition of samples in G×I research is the potential confounding influence of population stratification. Systematic variation in allele frequency among different ancestry groups can contribute to inaccurate associations between genes and an outcome of interest (Hellwege et al., Reference Hellwege, Keaton, Giri, Gao, Velez Edwards and Edwards2017). PCoA, PCA, and MDS are commonly used approaches to address population stratification and admixture in genetic analyses (Price et al., Reference Price, Patterson, Plenge, Weinblatt, Shadick and Reich2006). Accounting for genetic ancestry principal components can reduce error and improve power to detect true genetic effects by minimizing the influence of genetic associations with ancestry (Reich, Price, & Patterson, Reference Reich, Price and Patterson2008). Large-scale genome-wide association studies with diverse samples would also greatly enhance the strength of G×I studies by providing discovery samples with appropriately matched ancestry to determine minor allele frequency or create polygenic scores. However, there are also emerging methods such as multi-ethnic polygenic risk scores, which leverage the diversity in the target sample along with weights from large discovery samples to improve genetic risk prediction in diverse groups (Coram, Fang, Candille, Assimes, & Tang, Reference Coram, Fang, Candille, Assimes and Tang2017; Márquez-Luna, Loh, & Price, Reference Márquez-Luna, Loh and Price2017). With genotyping growing more accessible, there is opportunity to capitalize on existing large-scale prevention and intervention trials for G×I research by re-contacting participants for DNA samples. This may prove to be a helpful method for increasing the racial/ethnic diversity in genetic research by targeting existing samples of underrepresented groups. Recruiting and including individuals of non-European ancestry in genetic research is critical to ensure that all groups benefit from G×I findings.
Ethical considerations
The complex ethical implications of incorporating genetics into prevention and intervention of alcohol and other substance use were discussed in several of the reviewed papers (Albert et al., Reference Albert, Belsky, Crowley, Latendresse, Aliev, Riley and Dodge2015; Schlomer et al., Reference Schlomer, Cleveland, Feinberg, Wolf, Greenberg, Spoth and Vandenbergh2017; Vandenbergh et al., Reference Vandenbergh, Schlomer, Cleveland, Schink, Hair, Feinberg and Redmond2015). Discoveries about the conditional effects of prevention/intervention for individuals with certain genetic profiles might lead to concerning developments in the allocation of health services or determination of insurance eligibility. For example, one might imagine a future in which genetic risk is treated as a pre-existing condition, such that individuals with high liability for certain costly health conditions might be declined insurance coverage. We also foresee the possibility that treatments could be denied if there is G×I research suggesting it is ineffective for individuals with certain genetic risk profiles. Children might be differentially enrolled in prevention programs based on the likelihood that they will respond, while others might be denied those resources. A researcher in the UK, in collaboration with the Wellcome Trust, recently announced a program that would selectively provide additional education services to children from disadvantaged families with the highest and lowest polygenic scores for educational attainment (Griffiths, Reference Griffiths2019). It is feasible to imagine that this type of selective programming might be extended to the healthcare setting. The Genetic Information Nondiscrimination Act (GINA) of 2008 protects against discrimination by health insurance companies and employers, but the scope of the law is also limited in some ways. For example, it currently does not apply to life insurance, disability insurance, or small businesses with fewer than 15 employees. Some states have taken further action to expand the scope of GINA, but there remains concern over the ways that new and emerging technology might open the door to discriminatory practices in the future. There is also potential for public misunderstanding of results from G×I research, the implications of which might be quite deleterious. Genetic literacy is low even among those with high levels of education (Chapman et al., Reference Chapman, Likhanov, Selita, Zakharov, Smith-Woolley and Kovas2019), and genetic essentialism, the notion that genetic effects are immutable and fully determine various outcomes, is prevalent (Dar-Nimrod & Heine, Reference Dar-Nimrod and Heine2011; Pearson & Liu-Thompkins, Reference Pearson and Liu-Thompkins2012).
The NIH has a dedicated extramural funding program for research on the ethical, legal, and social implications (ELSI) of genomics, and also recently released a notice of special interest in research focusing on bioethical issues. Some findings from ELSI research suggest that providing genetic feedback on psychiatric conditions may lead to positive effects, such as reducing stigma, improving knowledge, increasing understanding of risk, and increasing patients’ self-report of empowerment (Austin, Reference Austin2019, Hippman et al., Reference Hippman, Ringrose, Inglis, Cheek, Albert, Remick and Honer2016, Kalb et al., Reference Kalb, Vincent, Herzog and Austin2017). Other studies indicate that genetic/biomedical explanations for psychiatric disorders may reduce belief in the effectiveness of certain treatment options and the likelihood of healing (Lebowitz, Reference Lebowitz2019). The degree to which this extends to perceptions surrounding effectiveness of prevention or early interventions among both parents and children is yet to be determined. In light of these questions and concerns, as a field we need to grapple with the challenging issues surrounding how genomics interfaces with prevention science.
We are likely to confront these issues sooner than many people might realize. Over the last several years, there has been an exponential increase in the public's interest in receiving personalized genetic information. More than 26 million individuals had received personalized genetic information by 2019 (Regalado, Reference Regalado2019). Free, publicly available websites allow individuals to upload raw genetic data obtained from these companies or from research studies to compute GPS for a variety of conditions. Psychiatric and substance use outcomes are some of the most accessed risk scores (Folkersen et al., Reference Folkersen, Pain, Ingasson, Werge, Lewis and Austin2019). These trends suggest that it will become increasingly common for individuals to have access to their genetic risk information in the future. Accordingly, understanding whether prevention and intervention programs vary in effectiveness for individuals at different levels of genetic vulnerability is likely to be useful and applicable information in the future. We are already seeing ways in which this information has been applied to other influential areas of human behavior, for example, by proposing to selectively enroll children with high and low polygenic scores for educational attainment into supplemental education services (Griffiths, Reference Griffiths2019). While we believe that based on our current limited understanding of genetic and environment influences on substance use behaviors, individuals should not be singled out for involvement in prevention/intervention based on genetic risk, we are faced with a future that is likely to make the integration of genetics into many realms currently unimaginable a reality. This creates an urgency to understand the ways environments moderate genetic risk for substance use problems, and what additional services are beneficial for at-risk individuals.
Limitations of the review
There are a few limitations of this systematic review that are important to acknowledge. First, the conclusions of the review are based on studies that met criteria for inclusion. In order to reduce the confounding effect of gene–environment correlation, we included only randomized studies in the review. Other studies, such as research on rates of relapse among treated individuals (e.g., Dahlgren et al., Reference Dahlgren, Wargelius, Berglund, Fahlke, Blennow, Zetterberg and Balldin2011; Wojnar et al., Reference Wojnar, Brower, Strobbe, Ilgen, Matsumoto, Nowosad and Burmeister2009), may provide additional insights and expand the scope of G×I research. Recovery from SUDs, particularly in the context of genetically-informed designs, is presently an understudied area of research. Additional research on recovery is warranted. Second, our review focused only on psychosocial intervention programs for alcohol and other substance use and excluded studies that incorporated a pharmacological component to treatment. Evidence suggests that treatment of psychiatric conditions is often most successful when a combination of pharmacological and psychosocial treatment is used (Anton et al., Reference Anton, O'Malley, Ciraulo, Cisler, Couper and Donovan2006; Balldin et al., Reference Balldin, Berglund, Borg, Månsson, Bendtsen, Franck and Willander2003; Dugosh et al., Reference Dugosh, Abraham, Seymour, McLoyd, Chalk and Festinger2016; Hien et al., Reference Hien, Levin, Ruglass, López-Castro, Papini, Hu and Herron2015). However, the inclusion of a “within the skin” treatment introduces complications in the context of understanding interactions between intervention and genetics on substance use outcomes. Nevertheless, understanding the interplay between genetic influences and pharmacological interventions is an important area of research and efforts to summarize findings of such studies are warranted.
Conclusions
The purpose of this review was to evaluate the current G×I literature focused on alcohol and other substance use behaviors, and discuss implications for future genetically-informed prevention and intervention research. G×I studies present a promising opportunity to improve prevention and intervention by increasing understanding of why individuals respond differentially to programs. Review of the 17 included studies suggests that the progression of G×I research on alcohol and other substance use has mirrored that of the broader genetics literature wherein early studies focused on candidate genes, and newer studies are beginning to incorporate genome-wide methods, such as genome-wide polygenic scores. All studies reported at least one significant G×I finding, which could be viewed as an indication that the effects of prevention programming frequently vary for individuals at varying genetic risk, though it also likely reflects publication bias (Duncan & Keller, Reference Duncan and Keller2011). Other themes that emerged from the literature include substantial heterogeneity in outcome measurement, a tendency to focus on youth with less attention to clinical and young adult populations, and limited representation of diverse populations. Most studies examined alcohol use, while other substances received less attention. Widespread and growing interest in obtaining information about one's own genotype is likely to shape health care and society in ways that we can only imagine right now. Research on how associations between genetic liability and behavioral outcomes vary as a function of prevention/intervention is likely to prove important as we approach a new age of precision medicine, one in which biological indicators are increasingly integrated into decisions about health promotion, prevention, and treatment.
Acknowledgments
This research was support in part by National Institute of Health grants F31AA027130 (PI: Neale) and K02AA018755 (PI: Dick). The authors declare no conflicts of interest.






