Children who experience chronic stress are at risk for problematic developmental outcomes including depression, post-traumatic stress disorder (PTSD), and cardiovascular and autoimmune diseases (Cicchetti & Toth, Reference Cicchetti and Toth2005; Melchior, Moffit, Milne, Poulton, & Caspi, Reference Melchior, Moffitt, Milne, Poulton and Caspi2007; Miller, Chen & Parker, Reference Miller, Chen and Parker2011). Stress experienced early in life may affect brain and behavioral development through biological embedding. Early-life experiences may alter biological processes which further influence developmental outcomes. Changes in DNA methylation could serve a mechanistic pathway through which biological embedding occurs. DNA methylation, or the addition of methyl groups to cytosines, is an epigenetic mechanism of gene regulation that is most often associated with gene silencing (Moore, Le, & Fan, Reference Moore, Le and Fan2013; Ziller et al., Reference Ziller, Gu, Müller, Donaghey, Tsai, Kohlbacher and Meissner2013), though this effect is dependent upon cytosine location in the genome (Cedar & Bergman, Reference Cedar and Bergman2009; Jones, Reference Jones2012; Maunakea, Chepelev, Cui, & Zhao, Reference Maunakea, Chepelev, Cui and Zhao2013).
Work in various rodent models and humans has associated developmental stress with long-term alterations in DNA methylation (Champagne et al., Reference Champagne, Weaver, Diorio, Dymov, Szyf and Meaney2006; Cicchetti, Hetzel, Rogosch, Handley, & Toth, Reference Cicchetti, Hetzel, Rogosch, Handley and Toth2016; Doherty, Foster, & Roth, Reference Doherty, Forster and Roth2016; Khulan et al., Reference Khulan, Manning, Dunbar, Seckl, Raikkonen, Eriksson and Drake2014; McGowan et al., Reference McGowan, Sasaki, D'Alessio, Dymov, Labonté, Szyf and Meaney2009, Naumova et al., Reference Naumova, Lee, Koposov, Szyf, Dozier and Grigorenko2012; O'Donnell et al., Reference O'Donnell, Chen, MacIsaac, McEwen, Nguyen, Beckmann and Meaney2018; Prados et al., Reference Prados, Stenz, Courtet, Prada, Nicastro, Adouan and Perroud2015; Roth, Lubin, Funk, & Sweatt, Reference Roth, Lubin, Funk and Sweatt2009; Unternaehrer et al., Reference Unternaehrer, Meyer, Burkhardt, Dempster, Staehli, Theill and Meinlschmidt2015; Weaver et al., Reference Weaver, Cervoni, Champagne, D'Alessio, Sharma, Seckl and Meaney2004, Yang et al., Reference Yang, Zhang, Ge, Weder, Douglas-Palumberi, Perepletchikova and Kaufman2013). In rodents, these epigenetic changes occur concurrently with behavioral alterations, such as altered stress responsivity and cognitive deficits (Doherty, Blaze, Keller, & Roth, Reference Doherty, Blaze, Keller and Roth2017; Weaver et al., Reference Weaver, Cervoni, Champagne, D'Alessio, Sharma, Seckl and Meaney2004). In humans, aberrant epigenetic alterations parallel many disorders and diseases, including depression, PTSD, and cardiovascular and autoimmune diseases (Dalton, Kolshus, & McLaughlin, Reference Dalton, Kolshus and McLoughlin2014; Yehuda et al., Reference Yehuda, Flory, Bierer, Henn-Haase, Lehrner, Desarnaud and Meaney2015; Handy, Castro, & Loscalzo, Reference Handy, Castro and Loscalzo2011; Farh et al., Reference Farh, Marson, Zhu, Kleinewietfeld, Housley, Beik and Bernstein2015).
Mounting evidence suggests that enhanced caregiving environments may alter epigenetic states. For example, cross-fostering rat offspring to high quality maternal care can reverse the methylation patterns that are associated with low quality maternal care or maltreatment (Roth et al., Reference Roth, Lubin, Funk and Sweatt2009; Weaver et al., Reference Weaver, Cervoni, Champagne, D'Alessio, Sharma, Seckl and Meaney2004). Among children, infant attachment, an indication of caregiving environment, is associated with variation in methylation of the genome (Garg et al., Reference Garg, Chen, Nguyen, Pokhivsneva, Chen and Unternaehrer2018). Further, a parenting intervention that is efficacious in reducing harsh parenting has been shown to protect children from the accelerated epigenetic aging that is associated with family risk (Brody, Yu, Chen, Beach, & Miller, Reference Brody, Yu, Chen, Beach and Miller2016). Parenting interventions that are delivered in early childhood appear to have persisting effects on the DNA methylome. Nurse Family Partnership (NFP) is an intervention that is delivered to first-time mothers who are at risk of child abuse. The mothers receive weekly visits from a community nurse beginning in pregnancy and continuing for approximately 30 months. When compared with a control condition, NFP is associated with numerous benefits to the parent–child dyad, including a reduction in child abuse and neglect, fewer behavioral and intellectual problems at age 6, and a reduction in child arrests at age 15 (Olds et al., Reference Olds, Eckenrode, Henderson, Kitzman, Powers, Cole and Luckey1997, Reference Olds, Henderson, Cole, Eckenrode, Kitzman, Luckey and Powers1998, & Reference Olds, Kitzman, Cole, Robinson, Sidora, Luckey and Holmberg2004). Among young adults whose mothers were enrolled in NFP when they were infants, variation in DNA methylation was associated with receipt of the intervention in early childhood, indicating a sustained impact of early intervention on the epigenome (O'Donnell et al., Reference O'Donnell, Chen, MacIsaac, McEwen, Nguyen, Beckmann and Meaney2018).
High-quality caregiving following maltreatment may change the epigenetic signatures associated with maltreatment. Following maltreatment, parental behavior can be improved through behavioral interventions that are delivered to the dyad (Bakermans-Krananburg, Van IJzendoorn, & Juffer, Reference Bakermans-Kranenburg, Van Ijzendoorn and Juffer2003; Toth, Gravener-Davis, Guild, & Cicchetti, Reference Toth, Gravener-Davis, Guild and Cicchetti2013). Dyadic interventions have been shown to be particularly efficacious in improving relationships and altering parent–child interactions (Toth et al., Reference Toth, Gravener-Davis, Guild and Cicchetti2013). The relational focus of dyadic interventions supports the child's development by altering continuous, dynamic interactions between the child and parent (Sameroff & Fiese, Reference Sameroff and Fiese2000). Attachment and Biobehavioral Catch-Up (ABC) is an intervention that is designed to enhance parenting among parents who are identified as being at risk for maltreating their children. The ABC intervention targets parental sensitivity and nurturance, with the objective of improving children's regulatory outcomes. The intervention has been shown to be efficacious at improving parenting quality in parents that are referred to Child Protective Services (Bick & Dozier, Reference Bick and Dozier2013), high-risk parents in a community setting (Caron, Weston-Lee, Haggerty, & Dozier, Reference Caron, Weston-Lee, Haggerty and Dozier2016), and parents that are adopting internationally (Yarger, Bernard, Caron, Wallin, & Dozier, Reference Yarger, Bernard, Caron, Wallin and Dozier2019). Furthermore, the parents that were randomized to receive ABC in the current sample were observed to be more sensitive in interactions with their children postintervention than those who were randomized to the control condition (Yarger, Hoye, & Dozier, Reference Yarger, Hoye and Dozier2016). The children whose parents received the ABC intervention were more likely to develop secure and organized attachments than those whose parents received a control intervention were (Bernard et al., Reference Bernard, Dozier, Bick, Lewis-Morrarty, Lindhiem and Carlson2012). Secure attachments in early infancy lead to positive developmental cascades; securely attached children are more likely to have positive peer relationships, higher self-worth, and fewer internalizing and externalizing problems than those who are insecurely attached (Weinfield, Sroufe, Byron, & Carlson, Reference Weinfield, Sroufe, Egeland, Carlson, Cassidy and Shaver2008). In addition to improving the quality of parent–child attachment, ABC improves biological functioning that is altered by stress. Children who participated in ABC were more likely to display normalized diurnal cortisol patterns than those whose parents received a control intervention (Bernard, Dozier, Bick, & Gordon, Reference Bernard, Dozier, Bick and Gordon2015). Preschool-aged children whose parents received ABC in infancy display stronger cognitive flexibility, greater theory of mind abilities, and they display less negative affect during a frustrating task than children whose parents received a control intervention (Lewis-Morriarty, Dozier, Bernard, Terracciano, & Moore, Reference Lewis-Morrarty, Dozier, Bernard, Terracciano and Moore2012; Lind, Bernard, Ross, & Dozier, Reference Lind, Bernard, Ross and Dozier2014). Moreover, changes in the hypothalamic-pituitary-adrenal (HPA) axis have been shown to persist for at least 3 years after having received the intervention (Bernard, Hostinar, & Dozier, Reference Bernard, Hostinar and Dozier2015). In middle childhood, children whose parents received ABC in infancy exhibited differential neural activity and autonomic nervous system functioning compared with those who received a control intervention (Bick, Palmwood, Zajac, Simons, & Dozier, Reference Bick, Palmwood, Zajac, Simons and Dozier2019; Tabachnick, Raby, Goldstein, Zajac, & Dozier, Reference Tabachnick, Raby, Goldstein, Zajac and Dozier2019). In summary, ABC is efficacious in altering children's attachment, socioemotional regulation, and biology.
Because epigenetic alterations may underlie the phenotypic outcomes that are associated with ABC, here, within a randomized controlled trial, we explored whether the ABC intervention is associated with changes in DNA methylation in young children who had experienced maltreatment. Based on prior literature demonstrating that in both human and nonhuman animal models early caregiving can change DNA methylation, we hypothesized that we would observe changes in DNA methylation within this sample due to the improvement in caregiving quality observed (Yarger et al., Reference Yarger, Hoye and Dozier2016). Furthermore, given the wide array of positive behavioral outcomes associated with ABC (e.g., improved HPA-axis functioning) and evidence documenting the effects of DNA on such outcomes (e.g., Bick et al,. Reference Bick, Naumova, Hunter, Barbot, Lee, Luthar and Grigorenko2012; Murgatroyd & Spengler, Reference Murgatroyd and Spengler2011), we also conducted exploratory analyses to identify the functional gene pathways that harbor differentially methylated CpG sites.
Method
In this study, 23 parent–child dyads with Child Protective Services (CPS) involvement as the result of allegations of maltreatment were randomly assigned to one of two interventions: the experimental intervention, ABC, or the control intervention, Developmental Education for Families (DEF). Upon parental consent, the children provided saliva samples for preintervention methylation analyses. The initial samples were collected in the children's homes following a 7-min period of free-play between child and parent. After the saliva sample was obtained, the children were randomized to ABC (n = 12) or DEF (n = 11) conditions. One month after completing the intervention, an additional research visit was conducted in the laboratory. Postintervention saliva samples were collected in the laboratory following the same 7-min free-play procedure between parent and child. Individuals who received DEF were not offered ABC after completing the intervention phase in order to allow for longitudinal assessment. All of the study procedures were approved by the Institutional Review Board at the University of Delaware.
Participants
The primary participants included 23 children who were involved with CPS. The parent–child dyads were referred to a local child welfare agency due to concerns of possible maltreatment. At the time of enrollment, the children ranged in age from 6 to 21 months (M = 14.07, SD = 4.18). The mothers were between the ages of 19.6 and 42.4 years old (M = 27.09, SD = 5.66). Demographic information did not differ between groups (see Table 1).
Table 1. Demographic characteristics by intervention group

Interventions
ABC is a 10-session manualized intervention that is designed to enhance parenting behaviors that are associated with effective self-regulation in children. The intervention was delivered in the participants’ homes by a certified parent coach. During the intervention, the parents are encouraged to provide nurturance, follow their children's lead with delight, and decrease frightening behaviors. Psychoeducation regarding the importance of these behaviors is provided through didactic content at each session. The parent coaches further reinforce desired parent–child interactions by making “in-the-moment” (ITM) comments that describe and label the parent behaviors and link these behaviors to associated outcomes. The frequency of in-the-moment comments predicts behavioral change among parents (Caron, Dozier, & Bernard, Reference Caron, Bernard and Dozier2016). The DEF intervention is structurally similar to ABC, with families participating in 10 weekly hour-long sessions within their homes. However, the interventionists provide psychoeducation and developmentally appropriate activities for parents to engage in with their child that are related to language development (e.g., labeling colors and shapes), motor skills (e.g., improving balance), and cognitive development (e.g., engaging in symbolic play) rather than parental sensitivity. The content of DEF was modeled after the daycare curriculum from the Carolina Abecedarian Project, but content regarding sensitive parenting was omitted in order to keep the interventions distinct (Ramey, McGinness, Cross, Collier, & Barrie-Blackley, Reference Ramey, McGinness, Cross, Collier, Barrie-Blackley and Borman1982).
DNA Methylation Measurement
Cotton swabs were used to collect saliva from the children (DNA Genotek, Ottowa, ON, Canada). Using saliva provides a minimally invasive collection procedure, and saliva methylation patterns often correlate positively with those in the blood and brain (Smith et al., Reference Smith, Kilaru, Klengel, Mercer, Bradley, Conneely and Binder2015; Thompson et al., Reference Thompson, Sharfi, Lee, Yrigollen, Naumova and Grigorenko2013). The saliva swabs were placed in Oragene discs (OGR-250) and stored at room temperature until extraction (DNA Genotek). The DNA was extracted with prepIT L2P following the manufacturer's instructions (DNA Genotek), with minor modifications. The modifications included centrifuging the samples at 14,000 rpm and rehydrating the dehydrated DNA pellet in 500 μL of TE buffer. Quality and quantity were verified by spectrophotometry (NanoDrop2000). The DNA samples were stored at -80°C until further analysis.
Bisulfite treatment, DNA amplification, sample hybridization to the Infinium HumanMethylation450 BeadChip, and measurement of the methylation values were performed by GenomeQuebec (Montréal, Canada). Methylation was measured using the Infinium HumanMethylation450 BeadChip array, which measures 485,000 CpG sites related to 99% of the genes that are included in the National Center for Biotechnology Information Reference Sequence Database (Illumina, San Diego, CA). The Infinium Methylation Assay provided a β-value for each CpG site, which indicates the proportion of methylated to unmethylated DNA that is present in the sample. The values ranged from 0, indicating a sample with no methylation present, to 1, indicating a fully methylated CpG site.
Analytic Plan
The methylation analyses were conducted in R (R Core Team, 2014). The raw values were normalized by using the ChAMP package (Morris et al., Reference Morris, Butcher, Feber, Teschendorff, Chakravarthy, Wojdacz and Beck2013). Sites were removed from further analyses if the probes had a detection rate of less than 0.01 (n = 2,023), aligned with known single nucleotide polymorphisms (n = 28,585), aligned with multiple locations (n = 8,489), or were present on the X or Y chromosome (n = 11,167). The data were further normalized via beta mixture quantile dilation, and the batch and chip effects were removed (Teschendorff et al., Reference Teschendorff, Marabita, Lechner, Bartlett, Tegner, Gomez-Cabrero and Beck2012). The differentially methylated CpG sites between groups were calculated in R, using empirical Bayes statistics for differential expression (Ritchie et al., Reference Ritchie, Phipson, Wu, Hu, Law, Shi and Smyth2015). To account for multiple comparisons, the p-values were adjusted with false discovery rate (FDR) Benjamini-Hochberg procedures. The primary analyses addressed whether the methylation sites differed as a function of the intervention. The methylation values for the ABC and DEF groups were compared before and after the interventions. The differentially methylated sites with an adjusted p-value < .05 were included in subsequent analyses.
The effects of potential covariates were accounted for by removing the sites that were associated with demographic variables (sex, minority status, and age) from the list of CpG sites that were used for further analyses. Due to our small sample size, racial categories were collapsed into “nonminority” and “minority” groups. The sites that were associated with sex (male and female) and minority status (minority and nonminority) groups were identified by using empirical Bayes statistics for differential expression (Ritchie et al., Reference Ritchie, Phipson, Wu, Hu, Law, Shi and Smyth2015). The sites that were associated with age were identified by regressing normalized β-values for each CpG site on child age. The CpG sites that were significantly associated (adjusted p-value < .05) with any demographic covariate (sex, minority status, or age) were removed (n = 755).
Secondary analyses were conducted to determine the likelihood of potential functional impact of changes in DNA methylation as a result of the intervention. Differentially methylated sites were first mapped to genic region, as the location of a CpG site within the gene has important implications for whether DNA methylation affects transcription. Next, we identified genes that contained several differentially methylated CpG sites. Next, we identified a subset of differentially methylated sites that were robustly associated with intervention status. A change in methylation score (Δβ) was calculated by subtracting the preintervention β from the postintervention β for each CpG site that differed between groups postintervention. Sites that changed in a consistent direction among all individuals in either group were selected and included in functional analyses that were performed in PANTHER version 11 (Mi, Muruganujan, Casagrande, & Thomas, Reference Mi, Muruganujan, Casagrande and Thomas2013; Mi et al., Reference Mi, Huang, Muruganujan, Tang, Mills, Kang and Thomas2017).
Results
Primary Analysis
As previously reported, the parents who received ABC exhibited a greater increase in parental sensitivity and decrease in intrusiveness than those who received DEF (Yarger, Hoye, & Dozier, Reference Yarger, Hoye and Dozier2016). Prior to beginning the intervention, 31,593 sites were differentially methylated between the ABC and DEF groups, but after FDR correction only one CpG site maintained statistical significance, t = −9.29, adjusted p < 0.01 (0.0017), cg09931793, Olfactory Receptor Family 2 Subfamily K Member 2 Gene (OR2K2). After the intervention, 82,937 sites were differentially methylated between the ABC and DEF groups, and after FDR correction 14,828 sites maintained their statistical significance (adjusted p < .05). Compared with the DEF group, 5,050 sites in the ABC group were hypermethylated and 9,778 were hypomethylated.
Secondary Analyses
The differentially methylated CpG sites that survived FDR correction were mapped onto gene regions (Table 2). Of the CpG sites that were associated with intervention groups, 42.16% fell within 1,500 base pairs of a transcription start site (TSS), or the location where transcription starts in a gene sequence, indicating that methylation changes at these sites are more likely to have functional relevance. Several genes contained multiple CpG sites that were differentially methylated between the groups, indicating that downstream processes are likely to be altered (Table 3). The gene functions that were associated with the list of genes were identified through Gene Ontology annotations (Ashburner et al., Reference Ashburner, Ball, Blake, Botstein, Butler, Cherry and Sherlock2000). While a large number of CpG sites were associated with intervention group differences, the functional relevance of these differences remains unknown. To identify the sites that are most likely to be associated with biological consequences, additional analyses were conducted. Selecting CpG sites with the largest between-group differences improves confidence in their functional relevance because larger between-group differences at a particular CpG site indicate more discrepant levels of methylation and, in theory, gene products, between the two intervention conditions. The average difference in β-values between groups was calculated for each CpG site. The top 10% of differentially methylated CpG sites (average difference in β > .025; n = 1,552) were used to identify the functional relevance of the intervention group differences through pathway analysis. Group differences were associated with 74 biological processes (Supplemental Table 1). Notably, the pathways that emerged as different between the two groups included cellular adhesion, cell signaling, neuronal development, signal transduction, and the regulation of metabolic processes (Table 4). The fold enrichment of each pathway indicates the degree to which pathway-associated genes appear among the 1,552 CpG sites relative to the expected frequency of sites associated with that pathway as estimated from the genome.
Table 2. Gene locations of CpG sites showing differential DNA methylation in association with the ABC intervention

Note: CpG sites may be aligned with more than one gene region.
Table 3. Genes containing 10 or more CpG sites associated with intervention group

Note: GO refers to gene ontology.
Table 4. Relevant biological pathways associated with intervention group differences in DNA methylation

Note: For the full table see Supplemental Table 1.
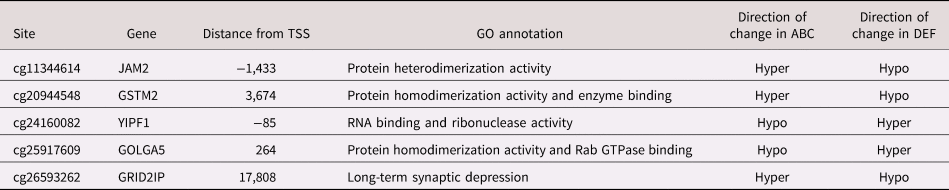
To identify robust changes in methylation associated with intervention, methylation patterns were investigated within each individual. The change in methylation scores (Δβ) was calculated for each individual at each of the 14,828 CpG sites that were associated with intervention group differences. The directionality of the changes in DNA methylation over time (preintervention to postintervention) was identified. Sites that changed in the same direction among all individuals within a group were identified. Within children who received DEF, methylation of 599 CpG sites changed in the same direction among all of the individuals within the group. Five CpG sites were associated with consistent changes in both groups, though the intervention groups changed in opposite directions (Table 5). The methylation values for the children who received ABC consistently changed in 181 CpG sites. To further investigate changes over time within the ABC group, the preintervention and postintervention methylation values were compared. The methylation values for 115 sites were significantly different before and after the intervention within the ABC group (Figure 1). The p-values were Bonferroni adjusted to account for multiple comparisons. The hierarchical cluster analysis of these sites is represented by dendrograms on the heatmap. Interestingly, all of the ABC postintervention cases clustered apart from both ABC pre- and DEF pre- and postcases, highlighting the potential influence of the ABC intervention on the DNA methylation profile. Whereas the majority of CpG sites postABC were hypomethylated in comparison to preABC, preDEF, and postDEF measurements, 13 CpG sites were hypermethylated. The 115 CpG sites aligned to 115 distinct genes. Of these sites, 55 were associated with promoter regions. Thirty-six CpG sites were located within 200 bp of the TSS, and 19 additional sites fell within 1500 bp of the TSS. Twenty CpG sites were located within the gene body, 12 sites were located within the 5′ UTR, and 10 sites fell within the 1st exon.

Figure 1. Heatmap representing DNA methylation levels of 115 robustly differentially methylated CpG sites. Each column represents an individual and each row represents one CpG site. The color gradient scale represents relative methylation values. Hierarchical clustering was performed with city block distance.
Table 5. CpG sites that changed consistently in all of the children in both ABC and DEF groups

Note: TSS refers to transcription start site. GO refers to gene ontology.
Discussion
In the current exploratory study, ABC was implemented with parent–child dyads that had been referred to child welfare because of concerns regarding maltreatment. Over the 10 sessions, parents who received ABC displayed increases in sensitive, attuned behaviors and decreases in intrusive interactions (Yarger et al., Reference Yarger, Hoye and Dozier2016). The ABC intervention was also associated with widespread changes in DNA methylation. Prior to receiving the intervention, children within the ABC and DEF groups on average had similar methylation levels after FDR correction. After the intervention, 14,828 CpG sites were differentially methylated in the two groups. The ABC intervention resulted in consistent changes in DNA methylation of 115 CpG sites among all of the individuals who received the intervention. Clustering of the children's DNA methylation by state of methylation of these sites separated the ABC intervention from all of the other groups, illustrating the specific influence of the ABC intervention on the DNA methylation profile. Children who received the control intervention displayed consistent changes in DNA methylation sites at 599 sites. These data suggest that children who continue to experience low parental sensitivity exhibit a methylation profile that worsens over time. While longitudinal studies of whole-genome methylation profiles in relation to early-life stress do not yet exist, these findings are consistent with literature regarding the effects of institutional care, which indicate that prolonged exposure to adverse environments are associated with increasingly aberrant brain functioning (Stamoulis, Vanderwert, Zeanah, Fox, & Nelson, Reference Stamoulis, Vanderwert, Zeanah, Fox and Nelson2015; Tottenham et al., Reference Tottenham, Hare, Quinn, McCarry, Nurse, Gilhooly and Thomas2010; Vanderwert, Marshall, Nelson, Zeanah, & Fox, Reference Vanderwert, Marshall, Nelson III, Zeanah and Fox2010). Overall, our data are consistent with findings among older children, suggesting that interventions that enhance parenting can alter children's DNA methylation patterns (Brody et al, Reference Brody, Yu, Chen, Beach and Miller2016; O'Donnell et al., Reference O'Donnell, Chen, MacIsaac, McEwen, Nguyen, Beckmann and Meaney2018).
Sites showing methylation changes mapped to various gene regions, all of which could have functional consequences on gene expression (Table 2; Brenet et al., Reference Brenet, Moh, Funk, Feierstein, Viale, Socci and Scandura2011; Cedar & Bergman, Reference Cedar and Bergman2009; Moore, Le, & Fan, Reference Moore, Le and Fan2013; Toperoff et al., Reference Toperoff, Aran, Kark, Rosenberg, Dubnikov, Nissan and Hellman2011). The CpG sites that were differentially methylated between the intervention groups aligned to 9,381 individual genes. Thirteen genes contained 10 or more differentially methylated CpG sites (Table 3). In determining the functional relevance of DNA methylation differences, genes that contain multiple differentially methylated sites are more likely to be affected, although a change in methylation at only a few sites within a gene is sufficient to significantly reduce the activity of that gene (Baker-Andersen, Ratnu, & Bredy, Reference Baker-Andresen, Ratnu and Bredy2013; Lubin, Roth, & Sweatt, Reference Lubin, Roth and Sweatt2008). The affected genes are involved in a wide range of cellular processes, including calcium ion binding, transcription factor binding, and DNA binding.
Pathway analyses were conducted using the CpG sites that had the greatest degree of differential methylation between children who had received ABC and those who had received DEF (Table 4). These sites are associated with numerous biological pathways, including the regulation of cell signaling, regulation of signal transduction, neuronal differentiation, regulation of a wide range of metabolic processes, and neuron development.
Though speculative, the functional analyses that were conducted within the present study are consistent with previous research on the outcomes that are associated with ABC. Attachment and Biobehavioral Catch-Up is associated with a wide variety of behavioral and biological outcomes. When compared with a control intervention, the children whose parents received ABC exhibited more normative patterns of diurnal cortisol production, neural activity, and autonomic nervous system functioning (Bernard et al., Reference Bernard, Dozier, Bick and Gordon2015; Bick et al., Reference Bick, Palmwood, Zajac, Simons and Dozier2019; Tabachnick et al., Reference Tabachnick, Raby, Goldstein, Zajac and Dozier2019). This broad array of outcomes suggests that fundamental biological functions, such as cell-signaling, cell adhesion, and neuronal development may be affected by the intervention. The present analyses of the biological pathways that are associated with differential DNA methylation suggest that ABC alters calcium ion signaling, neurogenesis, cell-substrate adhesion and metabolism. These biological functions may have broad effects on various types of biological regulation and behavior, such as those that were observed as outcomes of ABC.
Limitations
This study has several limitations that should be considered. First, these results should be considered preliminary because this was an exploratory study of methylation within a randomized controlled trial with no replication sample. Second, the present study was limited by a small sample size. However, using a small sample size for whole genome analyses may provide information regarding candidate genes to be analyzed in a large sample using more cost-effective methods. Though it is plausible that the ABC intervention caused beneficial changes in DNA methylation and/or avoided detrimental changes in DNA methylation that are associated with continued exposure to maltreatment, the data presented here should be interpreted with caution and viewed as groundwork for the generation of several hypotheses to be tested in future studies that explore the malleability of epigenetic states that are associated with maltreatment. Specifically, future research may consider whether a similar, consistent epigenetic signature of maltreatment is observed among a cohort of young children. Moreover, future studies may investigate whether such a signature persists over time, is affected by the quality of parenting, or is associated with various outcomes that are typically associated with early experiences of stress, such as biological dysregulation and deficits in impulse control. Lastly, DNA methylation was examined in saliva samples, which contain various proportions of epithelial cells, leukocytes, and bacteria. As methylation values vary by tissue type, differences in cell-type proportions may affect the measurement of methylation (Smith et al., Reference Smith, Kilaru, Klengel, Mercer, Bradley, Conneely and Binder2015). Further, we were unable to directly assess methylation within the brain, the organ that is most relevant to the behavioral intervention. It is obviously impossible to measure DNA methylation in the brain in living individuals, and several studies have shown strong correlations between peripheral and central measures of DNA methylation (Smith et al., Reference Smith, Kilaru, Klengel, Mercer, Bradley, Conneely and Binder2015; Thompson et al., Reference Thompson, Sharfi, Lee, Yrigollen, Naumova and Grigorenko2013). Consistent with previous studies, we discovered informative DNA methylation associations with a behavioral intervention in a peripheral tissue (Brody et al., Reference Brody, Yu, Chen, Beach and Miller2016; O'Donnell et al., Reference O'Donnell, Chen, MacIsaac, McEwen, Nguyen, Beckmann and Meaney2018; Perroud et al., Reference Perroud, Salzmann, Prada, Nicastro, Hoeppli, Furrer and Malafosse2013; Roberts et al., Reference Roberts, Lester, Hudson, Rapee, Creswell, Cooper and Eley2014). Interestingly, many of the pathways that were identified in the present study involve neuronal pathways. These data further support the relevance of peripheral DNA methylation measures to behavioral phenotypes.
Conclusions
In summary, this study is one of the first to explore parenting intervention effects on children's DNA methylation in this young age group, with results suggesting that enhancing parental behaviors during a child's early life changes DNA methylation. Importantly, these changes occurred in a short time span, indicating that an early intervention caused rapid alterations to the methylome. These findings warrant further large-scale research, including identifying whether changes are long-lived and are associated with positive behavioral change in children. Our data suggest several specific CpG sites and genes that may be influenced by early adversity and early intervention, providing a framework for candidate-gene approaches in larger cohorts. Nonetheless, these data suggest the remarkable malleability of a child's epigenome and the potential reversibility of epigenetic states that are associated with maltreatment, contributing to a growing body of research that seeks to understand whether DNA methylation could play a meaningful role in linking early-life experiences to physical and mental health.
Supplementary Material
To view supplementary material for this article, please visit https://doi.org/10.1017/S0954579419001421
Acknowledgments
This work was supported by donor funds from Edna Bennett Pierce to M. Dozier.