There is a burgeoning consensus among scholars that depressive symptoms follow a normative, inverted U-shaped trajectory before and during the transition to adulthood, which peaks in late adolescence and falls in young adulthood (e.g., Adkins, Wang, Dupre, van den Oord, & Elder, Reference Adkins, Wang, Dupre, van den Oord and Elder2009; Ge, Natsuaki, & Conger, Reference Ge, Natsuaki and Conger2006). Further, research has also consistently shown significant between-individual variation around mean trajectories (Adkins et al., Reference Adkins, Wang, Dupre, van den Oord and Elder2009; Adkins, Wang, & Elder, Reference Adkins, Wang, Elder, Turner and Schieman2008). Explaining individual differences in adolescent and young adult depressive symptom trajectories has proven a difficult task, with well-specified models including exhaustive lists of social risk factors explaining only modest amounts of trajectory variance (Adkins et al., Reference Adkins, Wang, Dupre, van den Oord and Elder2009; Natsuaki, Biehl, & Ge, Reference Natsuaki, Biehl and Ge2009). This has led to growing interest in the role of genetics in explaining individual differences in the development of depressed affect, with experts increasingly drawing on the diathesis-stress perspective to empirically investigate Gene × Environment (G × E) interaction (e.g., Caspi et al., Reference Caspi, McClay, Moffitt, Mill, Martin and Craig2002; Costello et al., Reference Costello, Pine, Hammen, March, Plotsky and Weissman2002).
This interest among behavioral scientists in the role of genetics in explaining developmental patterns of depressed affect is supported by several lines of inquiry within genetics. Although it has long been known that depression is substantially heritable (Sullivan, Neale, & Kendler, Reference Sullivan, Neale and Kendler2000), recent research has indicated that genetic influences on affect may vary considerably across development. For instance, biometric genetics research has shown that the heritability of depression significantly varies across adolescence and young adulthood, suggesting that the influence of various genes may increase or decrease across this important developmental period (e.g., Bergen, Gardner, & Kendler, Reference Bergen, Gardner and Kendler2007). Moreover, some research in this vein has indicated that distinct sets of genetic factors contribute to depressed affect at different points in development (Scourfield et al., Reference Scourfield, Rice, Thapar, Harold, Martin and McGuffin2003; Silberg et al., Reference Silberg, Pickles, Rutter, Hewitt, Simonoff and Maes1999). Reiss and Neiderhiser (Reference Reiss and Neiderhiser2000) have synthesized research in the area, presenting evidence of both quantitative and qualitative changes in genetic influence across development, while also arguing for the importance of environmental factors in moderating these changes. This perspective has proven prescient, receiving support from recent epigenetics research showing substantial gene expression changes across childhood and adolescence as developmental mechanisms “turn various genes off and on” (Whitelaw & Whitelaw, Reference Whitelaw and Whitelaw2006). Thus, beyond suggesting consistent gene effects across adolescence and young adulthood, contemporary genetics research has indicated that the influence of specific genetic loci may vary over the period.
Given this knowledge, it is perhaps surprising that virtually no research has considered Gene × Age interaction effects for candidate genes on depression trajectories across this developmentally dynamic life stage. The current study addresses this gap in the literature by investigating Gene × Age interaction on depressive symptom trajectories for five leading monoaminergic candidate genes, serotonin transporter linked polymorphic region (5-HTTLPR), dopamine receptor D4 (DRD4), monoamine oxidase A (MAOA), DRD2, and dopamine transporter 1 (DAT1), using false discovery rate (FDR) methods to control for the risks of false discoveries due to multiple testing. Below we discuss major streams of thought motivating this inquiry, including literature on depressive symptom trajectories, conventional static approaches on genetic influences on depressed affect, and emergent perspectives on developmental dynamism in genetic influences.
Depressive Symptom Trajectories Across Adolescence and Young Adulthood
Although longitudinal analyses of nationally representative data across adolescence and young adulthood remain uncommon, there is mounting evidence of a normative, inverted U-shaped pattern of depressed affect across this life course period. This conclusion is supported by longitudinal research finding curvilinear trajectories in samples of individuals moving through adolescence and young adulthood, as well as by research in younger samples showing linear increase through middle adolescence and studies of young adult samples showing linear decrease or stability through the twenties. For instance, inverted U-shaped trajectories have been found across ages 12–26 in former, methodologically robust analyses of the National Longitudinal Study of Adolescent Health (Add Health; Adkins et al., Reference Adkins, Wang, Dupre, van den Oord and Elder2009; Natsuaki et al., Reference Natsuaki, Biehl and Ge2009). Similarly, analyzing eleven waves of longitudinal data covering ages 12–23, Ge and colleagues (2006) found curvilinear trajectories of depressive symptoms, rising in early and middle adolescence and declining in late adolescence. Furthermore, Wight, Sepulveda, and Aneshensel (Reference Wight, Sepulveda and Aneshensel2004) examined depressive symptoms in three datasets (one adolescent sample and two adult samples) and found increasing levels in the adolescent sample, whereas the adult samples showed both lower initial levels and a steady decline over time. Consistent findings have been reported in several other analyses (e.g., Ge, Lorenz, Conger, Elder, & Simons, Reference Ge, Lorenz, Conger, Elder and Simons1994; Hankin et al., Reference Hankin, Abramson, Moffitt, Silva, McGee and Angell1998; Wade, Cairney, & Pevalin, Reference Wade, Cairney and Pevalin2002), collectively offering strong support for a normative curvilinear depressive symptom trajectory across this developmental period.
In addition to elucidating average trajectories of depressive symptoms in early life, research has also highlighted the longstanding issue of individual differences in the development of depression and depressive symptoms. For instance, recent trajectory analyses of Add Health using mixed effects modeling (Adkins et al., Reference Adkins, Wang, Elder, Turner and Schieman2008; Natsuaki et al., Reference Natsuaki, Biehl and Ge2009) and latent trajectory modeling (Adkins et al., Reference Adkins, Wang, Dupre, van den Oord and Elder2009) have shown that both intercept and slope trajectory components vary significantly across individuals, showing the majority of variance in the depressive symptoms measure comprises individual differences in these trajectory components. Although some of this variation may eventually be explained by improved measurement and modeling of social influences, there is a growing recognition that, as posited by the diathesis–stress model, a substantial portion of it is likely due to genetic factors (Caspi et al., Reference Caspi, McClay, Moffitt, Mill, Martin and Craig2002; Costello et al., Reference Costello, Pine, Hammen, March, Plotsky and Weissman2002).
Genetic Factors in Depression and Depressive Symptoms
Epidemiological research has offered strong evidence of the importance of genetics, with family studies indicating first-degree relatives of depressed probands to be 2.84 times more likely to experience major depression than controls, and twin studies indicating the heritability of unipolar depression to be 31% to 42% (Sullivan et al., Reference Sullivan, Neale and Kendler2000). However, despite the longstanding body of biometric genetics research showing substantial genetic influence, advances in mapping the molecular underpinnings of the phenotype have been slow. Although no consensus has been reached regarding the primary molecular mechanisms underlying mood disorder susceptibility, a confluence of neurobiological, pharmacological, and molecular genetic evidence has supported an important role for monoaminergic neurotransmission, particularly the serotonergic and dopaminergic systems. Among the many candidate gene variants influencing these systems, polymorphisms in 5-HTTLPR, DRD4, MAOA, DAT1, and DRD2 are among the most promising.
Serotonin transporter (5-HTTLPR, locus symbol SLC6A4)
Among neurotransmission systems the serotonergic system has received the most attention for its involvement in several processes including brain development and synaptic plasticity. Located at 17q11.2, the serotonin transporter gene (5-HTT) encodes a protein critically involved in the control of serotonin (5-HT) function. Allelic variation in the transcriptional region of 5-HTT, known as 5-HTTLPR, has been associated with personality traits including anxiety and aggressiveness (Anguelova, Benkelfat, & Turecki, Reference Anguelova, Benkelfat and Turecki2003). Short and long 5-HTTLPR variants differentially influence transcription activity of the 5-HTT gene promoter and the consequent 5-HT uptake in lymphoblastoid cells. Although results of main effects of 5-HTTLPR on depression have been mixed (Anguelova et al., Reference Anguelova, Benkelfat and Turecki2003), Caspi et al. (Reference Caspi, Sugden, Moffitt, Taylor, Craig and Harrington2003) have drawn together several lines of experimental genetic research to theorize that 5-HTTLPR may moderate the serotonergic response to stress. Investigating this hypothesis, Caspi and colleagues (2003) found individuals possessing the short allele of 5-HTTLPR to present more depression in response to stressful life events (SLEs) than individuals homozygous for the long allele. Since this study, several studies have attempted replication, yielding both positive (e.g., Wilhelm et al., Reference Wilhelm, Mitchell, Niven, Finch, Wedgwood and Scimone2006) and null results (e.g., Surtees et al., Reference Surtees, Wainwright, Willis-Owen, Luben, Day and Flint2006).
DRD4
The DRD4 gene maps 11p15.5 and contains a functional variable number tandem repeats (VNTR) polymorphism in its third exon (Van Tol et al., Reference Van Tol, Wu, Guan, Ohara, Bunzow and Civelli1992). The variant repeats 2 to 11 times, with 2 (D4.2), 4 (D4.4), and 7 (D4.7) repeats being the most common alleles (Van Tol et al., Reference Van Tol, Wu, Guan, Ohara, Bunzow and Civelli1992). DRD4 shows high levels of expression in frontal area of the brain and the nucleus acumbens, which are areas associated with affective behaviors (Emilien, Maloteaux, Geurts, Hoogenberg, & Cragg Reference Emilien, Maloteaux, Geurts, Hoogenberg and Cragg1999; Oak, Oldenhof, & Van Tol Reference Oak, Oldenhof and Van Tol2000). In vitro studies have indicated that alleles with decreased affinity for dopamine (e.g., DRD4.7 and DRD4.2), transmit weaker intracellular signals in comparison with other DRD4 alleles, thus may promote depressed affect through suboptimal functionality (Asghari et al., Reference Asghari, Sanyal, Buchwaldt, Paterson, Jovanovic and Van Tol1995). Although several lines of research have suggested DRD4 as a candidate gene for mood disorders, association results have been mixed. Significant associations have been reported for depressive disorders (e.g., Manki et al., Reference Manki, Kanba, Muramatsu, Higuchi, Suzuki and Matsushita1996; Muglia et al., Reference Muglia, Petronis, Mundo, Lander, Cate and Kennedy2002), but other studies have failed to confirm these findings (e.g., Bocchetta, Piccardi, Palmas, Oi, & Del Zompo, Reference Bocchetta, Piccardi, Palmas, Oi and Del Zompo1999; Serretti et al., Reference Serretti, Cristina, Lilli, Cusin, Lattuada and Lorenzi2002). It has been suggested that these failures to replicate may have been due to underpowered samples (Lohmueller, Pearce, Pike, Lander, & Hirschhorn, Reference Lohmueller, Pearce, Pike, Lander and Hirschhorn2003), a view supported by a recent, comprehensive meta-analysis that found a strong significant association between the DRD4.2 allele and unipolar depression (Lopez et al., Reference Lopez, Croes, Sayed-Tabatabaei, Claes, Van Broeckhoven and van Duijn2005).
MAOA-uVNTR
Located on the short arm of the X chromosome (Xp11.23) (Sabol, Hu, & Hamer, Reference Sabol, Hu and Hamer1998), the MAOA gene is considered a likely depression candidate gene based on two lines of evidence. First, MAOA has a central role in controlling amine disposability at the synaptic cleft, preferentially metabolizes serotonin and norepinephrine (Bach et al., Reference Bach, Lan, Johnson, Abell, Bembenek and Kwan1988). Second, MAOA inhibitors have been found effective in the treatment of depression (Murphy, Mitchell, & Potter, Reference Murphy, Mitchell, Potter, Bloom and Bloom1994). Although several different polymorphisms in the MAOA gene have been identified, only the VNTR polymorphism has been shown to affect the transcriptional activity of the MAOA gene promoter. This polymorphic region consists of a 30-bp repeated sequence present in two, three, 3.5, four, or five copies. Alleles with 3.5 or four copies are transcribed 2 to 10 times more efficiently than those with two, three, or five copies of the repeat (Sabol et al., Reference Sabol, Hu and Hamer1998). Although this promoter VNTR has shown association with several affective disorders including recurrent major depression (Preisig et al., Reference Preisig, Bellivier, Fenton, Baud, Berney and Courtet2000; Schulze et al., Reference Schulze, Muller, Krauss, Scherk, Ohlraun and Syagailo2000), other studies have reported null results (e.g., Kunugi et al., Reference Kunugi, Ishida, Kato, Tatsumi, Sakai and Hattori1999).
DAT1 (locus symbol: SLC6A3)
The dopamine transporter gene DAT1 maps to chromosome 5p15.3 and has a functional VNTR with alleles ranging from 3 to 11 repeats (Vandenbergh et al., Reference Vandenbergh, Persico, Hawkins, Griffin, Li and Jabs1992). In the central nervous system, the dopamine transporter protein DAT mediates reuptake of dopamine from the synaptic cleft, and thus is largely responsible for the intensity and duration of dopaminergic neurotransmission (Storch, Ludolph, & Schwarz, Reference Storch, Ludolph and Schwarz2004). Given the central role of the dopaminergic system in neurobiological theories of depression, DAT1 represents a plausible depression candidate, with several lines of evidence linking it to affect. For instance, pharmacological animal studies have demonstrated that drugs affecting DAT function (e.g., cocaine and amphetamine) enhance dopaminergic signaling, which induces hyperactivity and other changes in mood and behavior. The protein's importance to normal behavior has been demonstrated in DAT knockout mice (Giros, Jaber, Jones, Wightman, & Caron, Reference Giros, Jaber, Jones, Wightman and Caron1996). Because of the lack of the transporter protein, these animals have constantly elevated dopaminergic neurotransmission resulting in hyperactive behavior and negating the effects of psychostimulants. The results from main effect genetic association studies of DAT1 to affective disorders have yielded mixed results (Kelsoe et al., Reference Kelsoe, Sadovnick, Kristbjarnarson, Bergesch, Mroczkowski-Parker and Drennan1996; Waldman, Robinson, & Feigon, Reference Waldman, Robinson and Feigon1997). However, DAT1 has recently been analyzed from a G × E perspective, with results indicating a significant interaction with maternal rejection on major depressive disorder onset and suicidal ideation (Haeffel et al., Reference Haeffel, Getchell, Koposov, Yrigollen, DeYoung and Klinteberg2008).
DRD2Footnote 1 (rs1800497)
The TaqI A single nucleotide polymorphism (SNP) rs1800497 is located 10-kb downstream of DRD2, in a protein-coding region of the adjacent ANKK1 gene (Fossella, Green & Fan, Reference Fossella, Green and Fan2006). This SNP was long thought to lie within DRD2 and is known to be relevant to dopaminergic function, predicting D2 receptor density (Noble & Cox, Reference Noble and Cox1997; Thompson et al., Reference Thompson, Thomas, Singleton, Piggott, Lloyd and Perry1997) and glucose metabolism in dopaminergic regions of the human brain (Noble, Gottschalk, Fallon, Ritchie, & Wu, Reference Noble, Gottschalk, Fallon, Ritchie and Wu1997). The association of the SNP to D2 function is generally thought to stem from linkage disequilibrium with functional variants within DRD2 (Neville, Johnstone & Walton, Reference Neville, Johnstone and Walton2004). Consequently, the polymorphism has been studied as a candidate for affective disorders, with studies generally focusing on the A1 minor allele as a risk variant, as functional studies of both humans and mice have shown individuals with the A1 allele to have lower density of dopamine D2 receptors throughout the brain (Noble & Cox, Reference Noble and Cox1997; Nobel et al., 1997). Empirical findings of direct affective influence have been mixed, however, with some studies finding significant associations (Li et al., Reference Li, Liu, Sham, Aitchison, Cai and Arranz1999) and others not (e.g., Serretti et al., Reference Serretti, Macciardi, Cusin, Lattuada, Souery and Lipp2000). Recent research has attempted to resolve this discrepancy using a G × E approach, finding a significant interaction between DRD2 and SLEs on depressive symptomatology (Elovainio et al., Reference Elovainio, Jokela, Kivimaki, Pulkki-Raback, Lehtimaki and Airla2007).
Age Moderation of Genetic Influence
The period of adolescence to young adulthood is among the most developmentally intensive periods in the life course. It is characterized by important biological changes, such as puberty, and also a dramatic shift in social environment as children's parent-dominated social experience gives way to an expanding range of social options. Moreover, these changes have been linked to variation in the influence of genetic factors in ways that are potentially relevant to Gene × Age interaction in depressive symptom trajectories. For instance, there is ample evidence of extensive gene expression changes during adolescence, during which genes may be de/silenced (i.e., “turned on and off”) through developmentally and environmentally induced epigenetic changes (Whitelaw & Whitelaw Reference Whitelaw and Whitelaw2006). Although puberty represents a particularly striking example of phenotypic change in response to developmental epigenetic change (Whitelaw & Whitelaw Reference Whitelaw and Whitelaw2006), both mouse and human studies have demonstrated that these epigenetic changes continue across young adulthood and, indeed, throughout the life course (Barbot, Dupressoir, Lazar, & Heidmann, Reference Barbot, Dupressoir, Lazar and Heidmann2002; Fraga et al., Reference Fraga, Ballestar, Paz, Ropero, Setien and Ballestart2005). Although no research has yet focused on epigenetic regulation of monoamine genes in adolescence and young adulthood, given the extensive epigenetic changes characterizing the period, it is plausible that these genes may be differentially expressed, suggesting a potential molecular mechanism for Gene × Age interaction in depressive symptom trajectories during the transition to adulthood.
Biometric studies offer another source of evidence indicating changes in the influence of genetics on depression across adolescence and young adulthood. Analyzing twin, family, and adoptee data, biometric genetic studies decompose phenotype variance into aggregate genetic and environmental components without reference to molecular data. Many of these studies have examined depression at various points in early life (e.g., Eley & Stevenson, Reference Eley and Stevenson1999; Silberg, Rutter, & Eaves, Reference Silberg, Rutter and Eaves2001), and some have modeled how aggregate genetic influence changes as a function of age (e.g., Nes, Roysamb, Reichborn-Kjennerud, Harris, & Tambs, Reference Nes, Roysamb, Reichborn-Kjennerud, Harris and Tambs2007). The results of this body of research are well summarized by a recent meta-analysis by Bergen et al. (Reference Bergen, Gardner and Kendler2007), who analyzed six studies with sample ages ranging from 8 to 28, showing that the heritability of depression significantly increases from approximately 21% at age 8 to 42% at age 28.
Bergen and colleagues (2007) offer two broad, nonmutually exclusive potential explanations for the increasing role of genetics in depression as individuals move through adolescence and young adulthood. They suggest that as individuals age out of childhood, the role of parental social control recedes and individuals begin to self-select into environments, allowing them to more readily express their genetic proclivities. For instance, with parents no longer structuring their time, college students with depressive tendencies may fail to maintain social ties and drift toward isolation. The authors also offer the possibility that developmental epigenetic changes may “turn on” novel genes, providing additional sources of genetic variance. Further, the two mechanisms may interact, with novel environmental exposures triggering epigenetic changes. Although these possibilities cannot be adjudicated between without longitudinal epigenetic data, they both provide convincing rationale for considering age variation in the effects of known depression candidate genes across adolescence and young adulthood.
Methods
Sample
Data were analyzed from three waves of the National Longitudinal Study of Adolescent Health (Add Health). Add Health is a large nationally representative, longitudinal sample of adolescents and young adults. The National Quality Education Database was used as the baseline sample frame, from which 80 high schools were selected with an additional 52 feeder middle schools. The overall response rate for the 134 participating schools was 79%. Of the over 90,000 students who completed in-school surveys during the 1994–1995 academic year, a sample of 20,745 adolescents in Grades 7–12 were selected and have been interviewed three times in 1994–1995, 1995–1996, and 2001–2002. A questionnaire was also administered to a selected residential parent of each adolescent. Further details of Add Health's sampling design, response rates, and data quality are well documented (http://www.cpc.unc.edu/projects/addhealth/design).
The current study analyzes the three waves of repeated measures data from the Add Health sibling subsample, for which DNA measures are available. The sibling sample is composed of groups of respondents residing in the same household, and includes individuals of various degrees of biological relatedness, ranging from monozygotic twins to unrelated individuals. Respondents were included in the analysis sample if they had nonmissing values on all variables on at least one assessment. The total analysis sample consisted of 5614 observations for 1909 individuals, with each individual contributing an average of 2.9 observations. Individuals were nested within 1129 households, with each household containing one to four individuals (1.7 on average).
Measures
Depressive symptoms
Depressive symptoms were measured using a 9-item scale derived from the conventional 20-item Center for Epidemiologic Studies Depression Scale (CES-D; Radloff, Reference Radloff1977). The 20-item CES-D is composed of questions on a number of physical and psychological symptoms of depression, which cluster into four factors: somatic, depressed affect, positive affect, and interpersonal relations (Ensel, Reference Ensel, Lin, Dean and Ensel1996; Radloff, Reference Radloff1977). The scale has been validated using confirmatory factor analysis (CFA) in adult samples of Whites and Blacks (Blazer, Landerman, Hays, Simonsick, & Saunders, Reference Blazer, Landerman, Hays, Simonsick and Saunders1998).Footnote 2 It has also been validated in samples of adolescents and young adults (Radloff, Reference Radloff1991). Fortunately, a 19-item CES-D was collected in the first two waves of Add Health and a comparison with the subscale (nine items) indicated a high correlation (r = .91 and .92 in Waves I and II, respectively). Individual items were coded on a four-point scale to indicate the frequency of symptoms occurring during the past week, ranging from never or rarely (0) to most or all of the time (3). The primary outcome used in this analysis is the simple average of the nine items.
In addition to using a simple average of the nine items available across all three survey waves, sensitivity analyses were conducted using (a) a CFA factor score of the nine items, (b) an average of the three depressed affect items collected in all waves, which have been shown to be measurement invariant across racial/ethnic and immigrant groups in Add Health (Perreira, Deeb-Sossa, Harris, & Bollen, Reference Perreira, Deeb-Sossa, Harris and Bollen2005), and (c) a factor score of these three items. It has been shown previously that the use of factor scores for phenotypic measurement refinement can improve power to detect genetic effects (e.g., van den Oord et al., Reference van den Oord, Kuo, Hartmann, Webb, Moller and Hettema2008). Further, analyses of the current data indicate that allowing factor loadings to vary significantly improves model fit for both the nine- and three-item measures. In addition to measurement invariance characteristics, the use of the three-item subscale was also indicated by both notably higher factor loadings for these items relative to the other six indicators, as well as stronger theoretical correspondence of the items to the depression construct (see Perreira et al., Reference Perreira, Deeb-Sossa, Harris and Bollen2005). Correlations were high between all four specifications of the depressive symptoms variable (r = .84–.98). A constant was added to the factor scores setting their minimum values equal 0, in order to increase comparability of model parameters across depressive symptom specifications.
Parental socioeconomic status
Add Health allows respondents to report parental education levels for resident mother and father figures. These variables describe the highest level of education that the parent has completed, and range from “never went to school” to “professional training beyond a four year college or university.” Based on previous analyses, these items were coded as continuous variables (Adkins et al., Reference Adkins, Wang, Elder, Turner and Schieman2008). For each respondent the mean was then taken of reported parental education levels, which improved the explanatory power of the variable relative to either parent's level singly. Household income was ascertained from the parental questionnaire and includes all sources of income from the previous year (measured in thousands of dollars), and was logged. Correlation between parental education and logged household income was moderate (r = .43), indicating collinearity was not problematically high. Socioeconomic status (SES) indicators were mean centered to aid in model interpretation.Footnote 3
SLEs
An additive index was used to measure cumulative exposure to stressful life events. Presented in Appendix A, the SLE index used here is derived from one developed by Ge and colleagues (1994). Established criteria for the development of the SLE index were used in modifying and expanding the measure for the Add Health survey (Turner & Wheaton, Reference Turner, Wheaton, Cohen, Kessler and Gordon1995). For instance, only acute events of sudden onset and of limited duration that occurred within 12 months of the interview were included (Turner & Wheaton, Reference Turner, Wheaton, Cohen, Kessler and Gordon1995). To ensure a complete coverage of stressful events, approximately 50 items from various domains of life (e.g., family, romantic and peer conflicts, academic problems, exposure to violence, death of family and friends) were included. A major challenge of operationalizing SLEs is longitudinal validity—as adolescents make the transition into adulthood, some stressors become irrelevant (e.g., expulsion from school) and other stressors become relevant (e.g., divorce). Thus, to ensure stress was appropriately measured at different life stages, slightly different set of items is used in Wave III to capture the different life experiences. An additive index was created from the selected items and is mean centered in the current analysis.
Social support
The social support index shown in Appendix B is a composite measure of perceived social support across Waves I and II. It assesses how the respondents feel about their relationship with their closest social ties including family, teachers and parents. A CFA of the items indicated adequate fit (comparative fit index [CFI] = 0.971; root mean square error analysis [RMSEA] = 0.06) when including wave-specific factors and item-specific correlated errors between the two waves. A simple average of all the social support items was calculated and mean centered in this analysis.
Race/ethnicity
Add Health allows respondents to indicate as many race and ethnic categories as deemed applicable. Approximately 4% of the participants report a multiracial/ethnic identity. Following criteria developed by Add Health data administrators, we assign one racial identity for persons reporting multiple backgrounds (http://www.cpc.unc.edu/projects/addhealth/data/using/code/race). This method combines Add Health's five dichotomous race variables and the Hispanic ethnicity variable as following: respondents identifying a single race are coded accordingly; respondents identifying as Hispanic were coded as such regardless of racial designation; those identifying as “Black or African American” and any other race were designated as Black; those identifying as Asian and any race other than Black were coded as Asian, those identifying as Native American and any race other than Black or Asian were coded as Native American, and those identifying only as “other” were coded as such.Footnote 4
Candidate genes
In Wave III in 2002, DNA samples were collected from a subset of the Add Health sample. Genomic DNA was isolated from buccal cells at the Institute for Behavioral Genetics, University of Colorado, using a modification of published methods (Freeman et al., Reference Freeman, Powell, Ball, Hill, Craig and Plomin1997; Lench, Stanier, & Williamson, Reference Lench, Stanier and Williamson1988; Meulenbelt, Droog, Trommelen, Boomsma, & Slagboom, Reference Meulenbelt, Droog, Trommelen, Boomsma and Slagboom1995). The average yield of DNA was 58 μg. All of the Wave III buccal DNA samples are of excellent quality and have been used to assess nearly 48,000 genotypes.
DAT1
The allelic distribution of the 40 base pair (bp) VNTR in the 3′ untranslated region of the gene has been determined in duplicate (two separate polymerase chain reaction [PCR] amplifications and analyses, 5224 genotypes). The allelic distributions in base pairs and number of repeats (#R) were 360 bp (7R), 0.29%; 400 bp (8R), 0.34%; 440 bp (9R), 21.67%; 480 bp (10R), 76.98%; and 520 bp (11R), 0.72%.
DRD4
The 48-bp VNTR element in the third exon was determined in duplicate as above (5224 genotypes). The allelic distributions were 379 bp (2R), 8.28%; 427 bp (3R), 3.06%; 475 bp (4R), 64.71%; 523 bp (5R), 1.45%; 571 bp (6R), 0.74%; 619 bp (7R), 20.63%; 667 bp (8R) 0.84%; 715 bp (9R), 0.08%; and 763 bp (10R), 0.19%.
SLC6A4
The 44-bp addition/deletion in the 5′ regulatory region was determined in duplicate as above (5224 genotypes). The allelic distributions were 484 bp (short allele), 42.11%; and 528 bp (long allele), 57.89%.
MAOA-uVNTR
The 30-bp VNTR in the promoter was determined in duplicate as above (5224 genotypes). The allelic distributions were 291 bp (2R), 1.26% (males), 1.19% (females); 321 bp (3R), 40.21% (males), 36.29% (females); 336 bp (3.5R), 1.04% (males), 1.14% (females); 351 bp (4R), 56.31% (males), 60.14% (females); 381 bp (5R), 1.18% (males), 1.24% (females).
DRD2 TaqIA
The polymorphic TaqI restriction endonuclease site was determined in duplicate as above (5224 genotypes). The allelic distributions were A2 (C), 74.06%, and A1 (T), 25.94%.
Analytical strategy
Add Health is typical among longitudinal datasets, in that it is organized by wave of assessment with variability in chronological age at each wave. However, given that developmental research has clearly demonstrated age to be a more meaningful time metric than wave for the study of depression trajectories (e.g., Ge et al., Reference Ge, Lorenz, Conger, Elder and Simons1994; Hankin et al., Reference Hankin, Abramson, Moffitt, Silva, McGee and Angell1998), the data have been restructured in this analysis to provide age-based measurements. Fortunately, the statistical method employed, linear mixed effects models, has been shown to effectively accommodate features of the restructured data, including unbalanced repeated measures, variable data schedules, and missing observations (Diggle & Kenward, Reference Diggle and Kenward1994; Willett, Singer, & Martin, Reference Willett, Singer and Martin1998).
Linear mixed effects models have long been established in the statistical literature for the analysis of clustered, nonindependent data (Searle, Reference Searle1971; Searle, Casella, & McCulloch, Reference Searle, Casella and McCulloch1992), and are known to be particularly advantageous for growth curve analyses of longitudinal data (Willett et al., Reference Willett, Singer and Martin1998). The following equation describes a simplified version of the general mixed regression model used to investigate age variation in the effects of the candidate genes on depressive symptoms (DS):
where j, i, and t index the three levels of data: sibling cluster (i.e., household), individual, and assessment, respectively. Thus, the model allows random effects at both the sibling cluster and individual levels. Conditional on the random intercepts µj0 and υji0 at the sibling cluster and individual levels, the siblings and repeated assessments are assumed to be independent. The household level random effect captures much of the influence of population stratification on the results. This is because it accounts for intercept variation in depressive symptoms between households, with the assumption that the household cluster should be a decent proxy for identical by descent genetic similarity. Further control of population stratification is gained by the inclusion of self-identified race/ethnicity in all models.
The base model, without genetic effects, controls for race/ethnicity, gender, age, age2, social support, parental education, household income, and SLEs.Footnote 5 This model is consistent with prevailing environmental theories of depression and has been empirically tested by the author in previously published analyses of Add Health (see Adkins et al., Reference Adkins, Wang, Elder, Turner and Schieman2008, Reference Adkins, Wang, Dupre, van den Oord and Elder2009). For the primary set of analyses, in addition to the base model, each estimated model included a genetic variable and interaction terms between the genetic variable and both age and age2, thus examining variation in genetic effects across age by modeling genetic effects on each of the three trajectory components: intercept, linear age slope, and quadratic age slope. Sensitivity analyses repeat this procedure for each of the three alternate specifications of depressive symptoms. After identifying the most promising candidate genes, the robustness of these models are tested in an additional sensitivity analysis, by square root transforming the CES-D and rerunning the models to eliminate the possibility that results are driven by outliers.
For 5-HTTLPR, DRD4, DAT1, and DRD2, analyses were conducted on the full sample of both males and females. An alternative approach was used for MAOA, as its location on the X chromosome complicates direct comparisons between males and females. This is because males have a single allele at this locus (as they have only a single X chromosome), making their characterization straightforward, whereas females have two alleles, one of which may be silenced to some degree via X-inactivation (Jansson et al., Reference Jansson, McCarthy, Sullivan, Dickman, Andersson and Oreland2005; Meyer-Lindenberg et al., Reference Meyer-Lindenberg, Buckholtz, Kolachana, Hariri, Pezawas and Blasi2006). Given this ambiguity, analyses of MAOA are stratified by gender, whereas the full sample is jointly analyzed for all other genes.
The case of MAOA in females is illustrative of a more pervasive issue: it is often unclear what the optimal specifications of allelic effects are. Examples of both additive effects, in which there is a dose–response relationship between number of the risk alleles and the phenotype, and dominance effects, where a single allele is sufficient to give the full phenotypic effect, abound in the psychiatric genetics literature. Moreover, in psychiatric genetics there are also documented instances in which heterozygosity at a given locus is associated with a greater or lesser phenotypic effect, compared to homozygotes of either allele (e.g., Chen, Rainnie, Greene, & Tonegawa, Reference Chen, Rainnie, Greene and Tonegawa1994; Guo, Roettger, & Shih, Reference Guo, Roettger and Shih2007). Although former human genetics research, animal studies, and functional analyses can be informative in selecting allelic effect specifications, this knowledge is incomplete at best, and expectations are frequently overturned. DRD4 is instructive in this regard: functional studies have generally implicated the 7R allele (Asghari et al., Reference Asghari, Sanyal, Buchwaldt, Paterson, Jovanovic and Van Tol1995), but a recent meta-analysis instead only showed significant association between the 2R allele and unipolar depression (Lopez et al., Reference Lopez, Croes, Sayed-Tabatabaei, Claes, Van Broeckhoven and van Duijn2005). The case of MAOA and delinquency is similarly instructive as Caspi and colleagues (2002) have reported G × E between the MAOA 3R and maltreatment, while Guo and colleagues (Guo, Roettger, & Cai, Reference Guo, Roettger and Cai2008) have instead shown evidence of both main effects and G × E with the 2R allele, offering no support for a role of the 3R allele in delinquency.
In short, the field of molecular genetics is not yet far enough advanced to definitively dictate how genetic variables are best specified in statistical tests of association. That is to say, the frequency of unexpected associations combined with relatively weak theory of genetic mechanisms suggests that approaches relying strictly on precedent to specify allelic effects are vulnerable to missing true associations. This line of logic recommends an empirical approach to systematically screen various allelic effect specifications and Gene × Age configurations. Moreover, in practice researchers conducting candidate gene studies often tacitly employ such empirical, exploratory methods, but do not adjust significance criteria to account for multiple testing (Colhoun, McKeigue, & Smith, Reference Colhoun, McKeigue and Smith2003). Indeed, the enormous problem of false discoveries in candidate gene research, with 19 out every 20 associations currently reported in the literature thought to be false, is largely due to researchers conducting multiple tests, but only reporting significant findings (Colhoun et al., Reference Colhoun, McKeigue and Smith2003; van den Oord, Reference van den Oord2008). Given these facts, experts have argued that optimal methods for genetic discovery should cast a wide net, using exhaustive exploratory techniques, yet explicitly recognize the reduced confidence in any single association and adjust significance criteria accordingly (van den Oord, Reference van den Oord2005, 2008). Research has indicated that controlling for the FDR is a superior method for achieving these aims in candidate gene studies with correlated tests, such as the current analysis (van den Oord, Reference van den Oord2005; van den Oord & Sullivan, Reference van den Oord and Sullivan2003).
FDR
For each allele of the five monoamine genes investigated in this study, additive, dominance, and heterogeneous allelic effects were tested, each in a separate mixed model. Thus, the primary analysis consisted of 69 models, one for each of the 69 allelic specifications tested (counting MAOA alleles separately for male and females).Footnote 6 In each of the 69 models of the primary analysis, there were three coefficients of substantive interest, the direct genetic effect and the Gene × Age and Gene × Age2 interaction effects, resulting in 207 coefficients of interest from the primary analysis. Clearly, tests evaluating the statistical significance of these 207 coefficients are not independent. Different allelic specifications of the same polymorphism are correlated and thus, their test statistics also exhibit correlation. The same is true for the direct genetic, Gene × Age and Gene × Age2 effects for the same allelic specification—their tests of association are not independent and should not be treated as such. The ability to handle such correlated tests is a key benefit of the FDR approach employed here (Fernando et al., Reference Fernando, Nettleton, Southey, Dekkers, Rothschild and Soller2004; Sabatti, Service, and Freimer, Reference Sabatti, Service and Freimer2003; Storey & Tibsharani Reference Storey and Tibshirani2003). Unlike traditional procedures for adjusting for multiple testing, such as Bonferroni correction, the FDR approach yields powerful and valid inference even at relatively high levels of correlation (Sabatti et al., Reference Sabatti, Service and Freimer2003; van den Oord, Reference van den Oord2005).Footnote 7
Standard p values for the genetic, Gene × Age, and Gene × Age2 coefficients from each estimated mixed model were concatenated and FDRs were estimated from the p value distributions. FDRs can be estimated in various ways and many standard statistical packages (e.g., R, SAS) have such estimation procedures implemented. The current study estimates an FDR for a chosen threshold p value t. If the m p values are denoted p i, i = 1, … , m, this can be done using the formula:
Thus, the FDR is estimated by dividing the estimated number of false discoveries (the number of tests times the probability t of rejecting a marker without effect) by the total number of significant markers (i.e., total number of p values smaller than t) that includes the false and true positives. To avoid arbitrary choices, each of the observed p values can be used as a threshold p value t. The resulting FDR statistics are then called q values. In the current analysis, associations with q < 0.1 are considered potentially interesting, indicating the 1 out of 10 reported findings would be expected to be a false discovery.Footnote 8 This procedure was repeated for each of the three sensitivity outcomes, producing 828 coefficients of interest in total. Data management and statistical analyses were conducted using Stata 11 (StataCorp LP; www.stata.com) and FDRs were calculated using R 2.10.0 (http://www.r-project.org/index.html).
Results
Figure 1 plots means for each of the four CES-D specifications examined, by age and gender. Notable patterns include elevated symptom counts in late adolescence for all CES-D specifications and both genders, with symptom counts peaking around age 18. Females exhibit substantially higher symptom levels than males across all ages for all outcomes. Lower symptom levels were observed for the three item and factor score CES-D specifications relative to the primary nine item outcome, indicating that depressed affect symptoms occurred less frequently than symptoms of other dimensions. All outcomes exhibited roughly the same over-time pattern.

Figure 1. Mean depressive symptom levels for four Center for Epidemiologic Studies Depression Scale specifications, plotted by age and gender.
Table 1 presents descriptive statistics for the environmental predictors at Wave I by gender. Several trends are evident. Demographically, the sample included slightly fewer males than females, and Add Health's oversample of minorities was apparent with all non-White racial/ethnic groups representing higher proportions of the sample than the national population. Respondents were primarily of high school age in Wave I, and both genders generally reported comparable levels of perceived social support. Measures of SES indicated that respondent's mean yearly household income was approximately $45,000–$50,000 and the mean parental educational attainment was approximately a high school degree. Finally, SLEs were more frequently reported by males (mean = 2.75) than females (mean = 1.85).
Table 1. Descriptive statistics and environmental predictors

Note: SLEs, stressful life events.
Table 2 shows the number of significant gene, Gene × Age, and Gene × Age2 effects at various q value (i.e., multiple testing adjusted p value) thresholds. Thus, the first row of Table 2 show that for the primary outcome, the nine item CES-D average, six coefficients were significant at q < 0.1 out of 207 coefficients tested. All significant results from the primary results were for models examining DRD4 in the full sample. The sensitivity analyses of alternate CES-D specifications indicated that out approximately 621 parameters tested, two effects were significant at q < 0.1. This indicates that there were a small number of effects with p values significantly lower than expected by chance given the number of tests, suggesting the presence of true effects.
Table 2. Number of significant candidate gene effects on depressive symptom trajectories at various q-value thresholds

Note: MAOA, monoamine oxidase A gene; CES-D, Center for Epidemiologic Studies Depression Scale.
Table 3 describes the strongest candidate gene effects on depressive symptom trajectories detected in the analysis (q < 0.1, referred to as “significant”). The first six and latter two rows describe significant findings for the primary and sensitivity outcomes, respectively. Findings from both sets of outcomes are sorted by p values in ascending order. All significant findings involved either DRD4 5R allele in the full sample or MAOA 3.5R genotype among males.
Table 3. Candidate gene effects on depressive symptom trajectories with q < 0.1

Note: CES-D, Center for Epidemiologic Studies Depression Scale; DRD4, dopamine receptor D4 gene; 5R, 5-repeat; MAOA, monomine oxidase A gene.
All significant findings in the full sample regard the DRD4 5R allele. Results indicate that individuals with the relatively uncommon 5R DRD4 allele (2.87%Footnote 9 of the full sample, n = 161) experience unique trajectories of depressive symptoms across the period, characterized by U-shaped depressive symptom development, with relatively high levels as preteens at baseline, declining through adolescence, and rising in young adulthood. As illustrated in Figure 2, this trajectory is roughly opposite the normative, inverted U-shaped pattern commonly seen across the period. This was found for various specifications of the DRD4 5R allele for the primary outcome.Footnote 10Table 4 shows all estimates from the DRD4 no 5R allele model for each of the four outcome specifications. The p values were lowest for the primary nine-item average CES-D specifications (p = .002 and .003 for Gene × Age and Gene × Age2 coefficients, respectively), but Gene × Age and Gene × Age2 interaction terms were also p < .01 for all three sensitivity CES-D specifications. Additional sensitivity analyses also supported the robustness of this finding. As shown in Appendix C, square root transformation of the CES-D to improve the normality of the distribution and reduce the influence of outliers substantially increased the significance of the parameters of interest (p < .001 for Gene × Age and Gene × Age2 coefficients for all four CES-D specifications). Overall, these results suggest, with a high degree of confidence, that individuals with the DRD4 5R genotype exhibit a unique trajectory, characterized by relatively low depressive symptom levels in adolescence and relatively high levels in early adulthood.
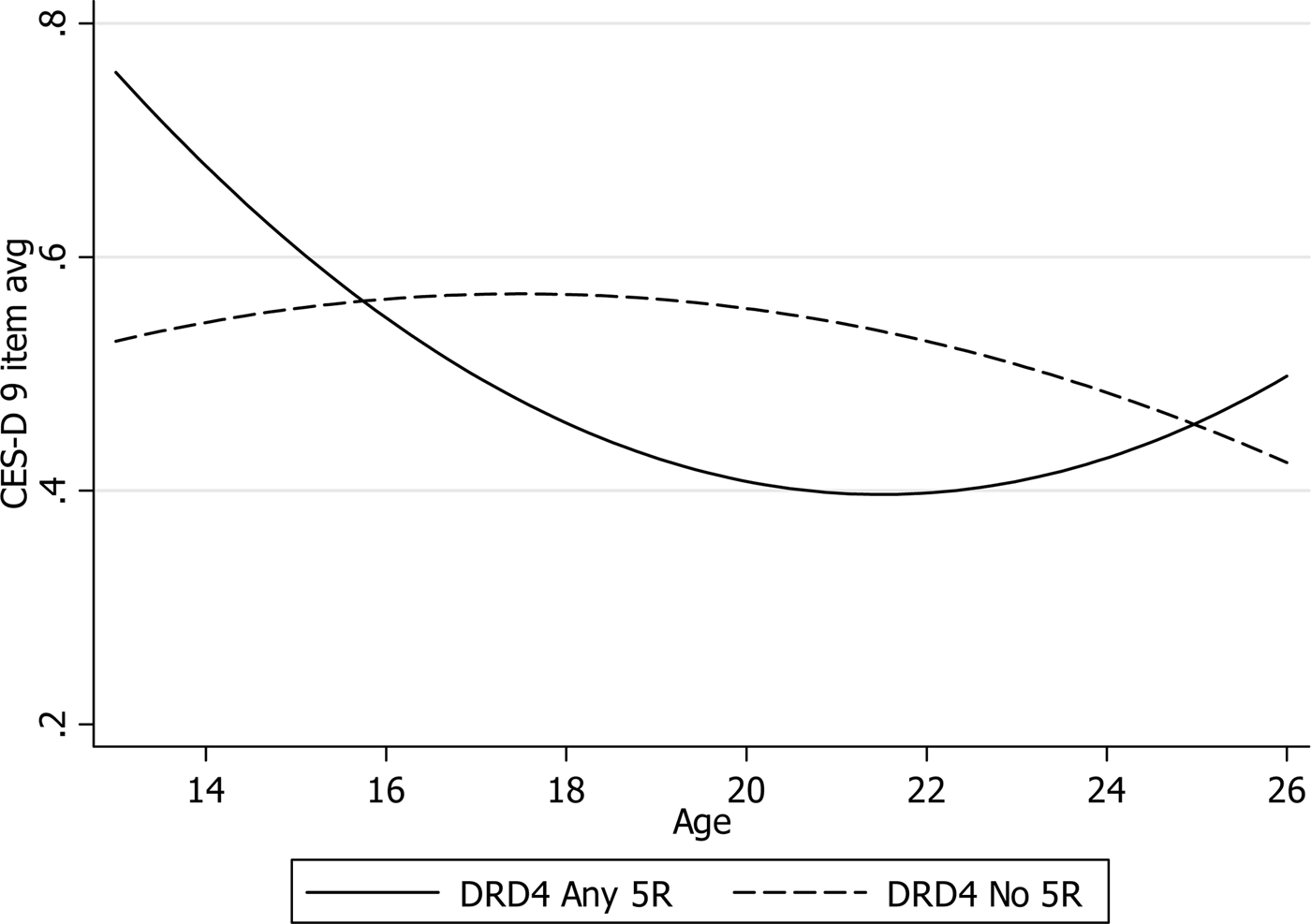
Figure 2. Depressive symptom age trajectory differences between dopamine receptor D4 5-repeat carriers and others.
Table 4. Parameter estimates of linear mixed models among full sample: Effects of DRD4 5R genotype on depressive symptom trajectories for 4 CES-D specifications

Note: The values in parentheses are p values. DRD4, dopamine receptor D4 gene; 5R, 5-repeat; CES-D, Center for Epidemiologic Studies Depression Scale; SLE, stressful life event.
*p < .05. **p < .01. ***p < .001.
The final significant finding regards the uncommon MAOA 3.5R genotype among males (1.04% of the male sample, n = 28). This allele was found to significantly interact with age and age2 in both the nine- and three-item CES-D factor score outcomes. As shown in Figure 3, compared to the normative pattern males with the 3.5R genotype, exhibited a similar, but markedly more curvilinear, inverted U-shaped trajectory. Although only the three-item factor score CES-D specifications satisfied the q < 0.1 threshold (p < .01 for three-item factor score Gene × Age and Gene × Age2 coefficients), as shown in Table 5, Gene × Age and Gene × Age2 interaction terms were p < .05 for all CES-D specifications. These results were largely supported by additional sensitivity analyses showing significant results for square root transformed specifications of the CES-D (Appendix D).Footnote 11 This finding suggests that males with the MAOA 3.5R genotype may experience a particularly distressful adolescence, before converging with their peers in early adulthood.

Figure 3. Depressive symptom age trajectory differences between male carriers of the monoamine oxidase A 3.5 genotype and other males.
Table 5. Parameter estimates of linear mixed models among male sample: Effects of MAOA 3.5R genotype on depressive symptom trajectories for 4 CES-D specifications
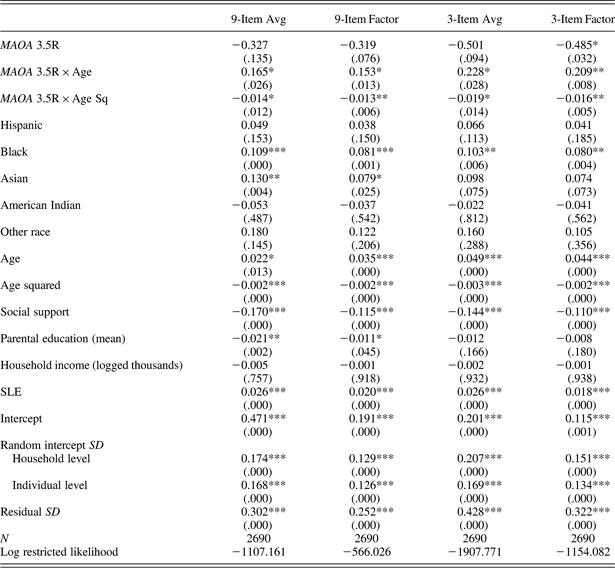
Note: The values in parentheses are p values. MAOA, monoamine oxidase A gene; 3.5R, 3.5-repeat; CES-D, Center for Epidemiologic Studies Depression Scale; SLE, stressful life event.
*p < .05. **p < .01. ***p < .001.
Discussion
Leading developmental perspectives have long stressed the importance of accounting for temporality and life course variation in models of mental health. A primary insight of such perspectives is that the importance of various depressogenic factors fluctuates across developmental trajectories (Elder, George, & Shanahan, Reference Elder, George, Shanahan, Kaplan and Kaplan1996; Willett et al., Reference Willett, Singer and Martin1998). The current study endeavors to wed this perspective to molecular genetic approaches to depressed affect. Although psychiatric molecular genetics has made advances toward elucidating the link between genetic variation and depression, virtually all of this research has been atemporal. The weakness of this static perspective on the genetic determinants of depression is highlighted not only by developmental perspectives, but also by newer research within genetics showing that epigenetic mechanisms “turn genes off and on” in response to developmental and environmental cues (Whitelaw & Whitelaw, Reference Whitelaw and Whitelaw2006). Using the Add Health genetic subsample, this study has addressed the issue of variation in genetic influences across adolescence and young adulthood through comprehensively testing the effects of five monoamine genes on depressive symptom trajectories, while employing FDR methods to control the risk of false discoveries.
The most promising associations detected were for interactions between the DRD4 dopamine receptor gene and age trajectory components in the full sample, and the MAOA VNTR promoter polymorphism and age trajectory components among males. Specifically, in the case of the DRD4 finding, individuals with the 5R allele were found to exhibit a roughly opposite trajectory compared to the normative inverted-U pattern. Thus, individuals with any 5R alleles were shown to have relatively low symptom levels through late adolescence, before experiencing increases in early adulthood. This pattern suggests that carriers of the DRD4 5R allele navigate their high school years with relative psychological ease compared to others, but begin to experience elevated distress as they transition into adult roles. Interpreting the molecular mechanism underpinning this finding is challenging, as very little is known about the 5R allele. Given its relatively low allele frequency (2.87% of the full sample), it has not been well characterized in functional studies; thus, its gene expression profile is poorly understood.
However, one potential explanation of the DRD4 5R finding stems from association studies linking DRD4 to substance abuse. The DRD4 5R allele has shown evidence of association to abuse of various substances, including alcohol (e.g., Muramatsu, Higuchi, Muramaya, Matsushita, & Hayashida, Reference Muramatsu, Higuchi, Murayama, Matsushita and Hayashida1996) and heroin (e.g., Li et al., Reference Li, Xu, Deng, Cai, Liu and Liu1997).Footnote 12 Although these findings remain controversial (see Lusher, Chandler, & Ball, Reference Lusher, Chandler and Ball2001), their potential relevance to the current DRD4 finding becomes apparent when considering the life course context of substance abuse. Specifically, social control factors limiting access and abuse of substances, such as parental monitoring and legal obstacles, are relatively strong in adolescence. In the late teens and early 20s, after individuals leave their parents' homes and can legally purchase alcohol, these social control mechanisms weaken and impediments to substance abuse are removed. Given that the upswing in depressive symptoms for DRD4 5R carriers observed here closely corresponds to the transition to adulthood, and that substance abuse and depression are highly correlated and frequently clinically comorbid (e.g., Grant & Harford, Reference Grant and Harford1995), it seems plausible that loosening social control may be a key explanatory factor of the elevated distress levels observed among 5R carriers in young adulthood. However, as the direction of causality between substance abuse and depression is debated and likely reciprocal to some degree (e.g., Aneshensel & Huba, Reference Aneshensel and Huba1983), future research will be needed to replicate this finding and disentangle the putative web of causality between DRD4, substance abuse, and depressive symptoms. Moreover, given the dearth of knowledge into the 5R allele's gene expression profile, basic molecular research will be necessary to validate the finding by characterizing the functionality of this uncommon variant.
The other notable substantive finding was an association between the MAOA 3.5R allele and depressive symptom trajectory components in the male sample. Specifically, males with the 3.5R genotype had more curvilinear symptom trajectories than the normative pattern, with higher peaks in late adolescence and sharper declines in early adulthood. Thus, males with the 3.5 genotype were shown to have a particular distressful time during high school and the subsequent transition to adulthood, but converge with their peers in early adulthood. This age variation in the influence of MAOA may explain inconsistencies in former MAOA–depression association results, which have shown both elevated depression levels among male carriers of the 3.5R and other long MAOA alleles (Du, Bakish, Ravindran, & Hrdina, Reference Du, Bakish, Ravindran and Hrdina2004; Yu et al., Reference Yu, Tsai, Hong, Chen, Chen and Yang2005), and also no significant association (Kunugi et al., Reference Kunugi, Ishida, Kato, Tatsumi, Sakai and Hattori1999). Furthermore, the current results may shed light on results from a recent meta-analysis of six MAOA–depression association studies, which found a strong trend toward increased depression among carriers of the 3.5R and other long MAOA alleles falling just short of statistical significance (odd ratio [OR] = 0.86; 95% confidence interval [CI] = 0.74–1.01; Lopez-Leon et al., Reference Lopez-Leon, Janssens, Ladd, Del-Favero, Claes and Oostra2008).Footnote 13 It is interesting that this meta-analysis found strong evidence of heterogeneity in effect sizes across studies. Results of the current study offer a potential explanation for this heterogeneity, suggesting that age differences across samples may be driving effect differences.
In sum, this study has shown that individual variation in adolescent and young adult depressive symptom trajectories is partially explained by specific genetic variants. These findings advance developmental perspectives through both addressing perennial issues and raising new questions. Depressive affect has long been of particular interest to developmental psychopathologists due to its multifactorial etiology, encompassing psychological, social, and biological factors (Cicchetti & Toth, Reference Cicchetti and Toth1998). In line with the tenants of this approach, we have explored the dynamic etiology of depressed affect by merging multiple levels of analysis and leveraging longitudinal trajectories to simultaneously examine the influence of specific genetic variants and environmental factors (Sroufe & Rutter, Reference Sroufe and Rutter1984; Zahn-Waxler, Klimes-Dougan, & Slattery, Reference Zahn-Waxler, Klimes-Dougan and Slattery2000). The results inform long-standing issues of central importance in the developmental literature. For instance, as Rutter & Sroufe (Reference Rutter and Sroufe2000) note, the increased frequency and intensity of depressed affect in adolescence is a primary feature of affective psychopathology and requires a developmental approach to elucidate its origin. Here we have shown that variation in monoaminergic genes strongly influences this pattern, predicting a particularly distressful adolescence for some (i.e., male MAOA 3.5R carriers), and relatively psychological ease in adolescence, followed by difficulty in young adulthood, for others (DRD4 5R carriers). Future developmental research would do well to continue examining the possibility that much of the adolescent elevation in depressed affect observed at the population level may be driven by genetically distinct subgroups.
This research also raises new questions germane to further developmental study. For instance, how do these polymorphisms exert affective influence? Given the temporal patterns observed, it is clear that some aspect of development moderates these genetic influences. It seems likely that fluctuations in gene expression levels, quite possibly epigenetically regulated, are involved (Bergen et al., Reference Bergen, Gardner and Kendler2007; Reiss & Neiderhiser, Reference Reiss and Neiderhiser2000), but what drives these expression/epigenetic changes? Is this a predominately biological phenomenon, similar, and perhaps related, to puberty (see Eaves et al., Reference Eaves, Silberg, Foley, Bulik, Maes and Erkanli2004)? Or do social environmental changes associated with adolescence influence gene expression levels for risk variants? Applying a developmental approach to examining correlations and interactions of social risk and protective factors to the implicated variants could yield empirical answers to these questions. Furthermore, these are not issues of purely academic interest. As persuasively argued by Reiss and Neiderhiser (Reference Reiss and Neiderhiser2000) the ability of social factors to buffer against genetic predispositions toward depressed affect has vital importance to intervention efforts. By more completely understanding the configurations of social and genetic factors contributing to depressed affect development, interventions can both identify genetic risk groups early on, and potentially modify environments to prevent psychopathological developmental cascades.
As is typical, this research is unlikely to be the final word on the topic. This investigation is limited by the number of waves of data, and therefore the trajectory length and age range, available. Additional waves of data, which are forthcoming, will allow an extension of our understanding of how depressive symptoms develop over a longer period of the life course. Future research could also benefit from increasing coverage of genetic variation. Although candidate gene approaches are apt to remain important in G × E studies into the near future, there is a progressive movement in genetics toward more exploratory analyses examining genetic variation across the genome. These genome-wide association studies typically include over 500,000 genetic markers, and although still relatively uncommon in behavioral research, the rapidly decreasing cost of genotyping guarantees that such data will soon become available for longitudinal, behavioral surveys. This development will represent a paradigm shift in G × E studies, allowing analysis of social moderation of genetic influences on an unprecedented scale. However, it will also pose challenges to behavioral scientists as they join statistical geneticists in grappling with how to best analyze such massive datasets. Although the FDR techniques employed here represent vanguard techniques for addressing the issues of multiple testing inherent to genome-wide association studies, this area will certainly remain an active research frontier into the foreseeable future.
Despite these limitations, the present study improves our understanding of depressive symptomatology in adolescence and young adulthood and advances a framework for future research in the area. Specifically, results show significant temporal variation in the effects of MAOA and DRD4 on depression. Beyond the substantive results, this study shows the value of combining temporally dynamic, developmental perspectives with comprehensive empirical statistical approaches to optimize the search for genetic influences across the life course. This can be seen from various aspects of the current study. First, without an exhaustive exploration of various allelic specifications beyond those conventionally assessed, highly significant associations for the 3.5R MAOA and 5R DRD4 alleles would not have been detected. Also, employing a developmental perspective to consider age variations in genetic influence enabled the detection of very strong nonlinear Gene × Age interactions that would have otherwise been missed. Finally, the use of FDR statistical methods allowed these comprehensive empirical explorations by controlling the risk of false discoveries, a major problem in genetic research (Colhoun et al., Reference Colhoun, McKeigue and Smith2003), that behavioral scientists interested in incorporating genetic perspectives have yet to sufficiently address.
Appendix A Items in Stressful Life Events Index

Appendix B Social Support Scale

Appendix C Parameter estimates of linear mixed models among full sample: Effects of DRD4 5R on square root transformed depressive symptom trajectories

Note: The values in parentheses are p values. DRD4, dopamine receptor D4 gene; 5R, 5-repeat; SLE, stressful life event.
*p < .05. **p < .01. ***p < .001.
Appendix D Parameter estimates of linear mixed models among males: Effects of MAOA 3.5R on square root transformed depressive symptom trajectories

Note: The values in parentheses are p values. MAOA, monoamine oxidase A gene; 3.5R, 3.5-repeat; SLE, stressful life event.
*p < .05. **p < .01. ***p < .001.














