Introduction
Communities of insects associated with plant galls are useful model systems for studying trophic interactions among insects (Csóka et al. Reference Csóka, Stone and Melika2005), and species level identification of the inhabitants is necessary to analyse food webs (Gómez et al. Reference Gómez, Nieves-Aldrey, Hernández Nieves and Stone2011). Besides the inducers, gall communities are commonly comprised of parasitoids that feed on inducers or other inhabitants, and inquilines that feed on galls tissues (Shorthouse Reference Shorthouse2010). One of the driving forces behind the speciation of gall parasitoids is the phenological patterns of their hosts, as parasitoids can only reach their hosts by oviposition through gall tissues, which proliferate and enlarge as the galls develop (reviewed in Csóka et al. Reference Csóka, Stone and Melika2005). Several studies have also shown that the host plant, host organ, and the time required for gall development all affect richness of parasitoid species (e.g., Schönrogge et al. Reference Schönrogge, Stone and Crawley1995, Reference Schönrogge, Stone and Crawley1996; Plantard et al. Reference Plantard, Rasplus and Hochberg1996; Plantard et al. Reference Plantard, Rasplus, Mondor, Le Clainche and Solignac1999); however, the reason why many parasitoids are highly host specific, despite the presence of other potential hosts on the same host plant, are largely unknown (Csóka et al. Reference Csóka, Stone and Melika2005). All known parasitoids that attack cynipid hosts are wasps within the superfamilies Chalcidoidea and Ichneumonoidea (Shorthouse Reference Shorthouse2010).
The superfamily Chalcidoidea contains an estimated 500 000 species, making it one of the most biologically and morphologically diverse groups of parasitic wasps (Gibson et al. Reference Gibson, Huber and Woolley1997, Reference Gibson, Heraty and Woolley1999; Munro et al. Reference Munro, Heraty, Burks, Hawks, Mottern and Cruaud2011). While some chalcidoids are phytophagous, the majority are entomophagous and their hosts include all life-history stages of 13 orders of insects, two orders of arachnids, and one family of nematodes (Gibson et al. Reference Gibson, Huber and Woolley1997, Reference Gibson, Heraty and Woolley1999). Chalcids within the family Eurytomidae have over 1400 nominal species in 84 genera and are found in most zoogeographical regions (Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007b; Gates Reference Gates2008; Noyes Reference Noyes2012). Eurytomids are largely endophytic as seed feeders, gall inducers or parasitoids of phytophagous insects (Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007b). The accurate identification of eurytomids, in particular the genus Eurytoma Illiger, which includes more than 700 described species, has proven difficult using existing morphological keys due to overlapping diagnostic characters and lack of illustrations (e.g., Bugbee Reference Bugbee1967). As a result, phylogenetic, ecological, and evolutionary studies of eurytomids have been impeded. The degree of morphological conservatism is particularly prominent in members of the Eurytoma rosae Nees species group, which parasitises various gall-inducing cynipids, Tephritidae (Diptera), and Curculionidae (Coleoptera) (Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007b). Members of the E. rosae group are often morphologically similar and impossible to segregate into morphospecies, despite being ecologically and genetically distinct (Ács et al. Reference Ács, Melika, Kalo and Kiss2002; Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007a; Gómez et al. Reference Gómez, Nieves-Aldrey, Hernández Nieves and Stone2011).
A total of 14 native species of cynipid wasps of the genus Diplolepis Geoffroy (Hymenoptera: Cynipidae) have been recorded in Canada, all of which induce structurally distinct galls on Rosa Linnaeus (Rosaceae) (Shorthouse Reference Shorthouse2010). These rose galls are heavily attacked by chalcid parasitoids, of which eurytomids are the most abundant (Shorthouse et al. Reference Shorthouse, Leggo, Sliva and Lalonde2005; Shorthouse Reference Shorthouse2010). Ten species of eurytomids are known to be associated with galls of Diplolepis in Canada feeding as koinobiont ectoparasitoids of either the inducers or cynipid inquilines of the genus Periclistus Förster (Noyes Reference Noyes2012). While most eurytomids are univoltine and overwinter within galls before exiting the following spring, cases of fall emergence have been recorded where mature eurytomids pupate and exit the gall in the fall of the year of gall initiation (Shorthouse Reference Shorthouse1973, Reference Shorthouse2010; Brooks and Shorthouse Reference Brooks and Shorthouse1997). Several studies have been conducted on both adult (Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007a, Reference Lotfalizadeh, Delvare and Rasplus2007b) and larval (Gómez et al. Reference Gómez, Nieves-Aldrey, Hernández Nieves and Stone2011) eurytomids associated with rose gall communities in the Palearctic region, whereas the Nearctic species have received little taxonomic attention. Bugbee (Reference Bugbee1951a, Reference Bugbee1951b, Reference Bugbee1973) described the majority of the Nearctic species; however, many of the original species descriptions are brief, and were often based on a limited number of specimens collected from a single locality. Further, morphological variations from the type specimens were assigned as subspecies, resulting in even more indistinguishable taxa. While identification keys were provided for Eurytoma by Bugbee (Reference Bugbee1951b, Reference Bugbee1967), they were based only on females and distinguishing characters used were often ambiguous and generally lacking illustrations. These impediments confound studies on host-parasitoid relationships within galls induced by Diplolepis (e.g., Shorthouse et al. Reference Shorthouse, Leggo, Sliva and Lalonde2005; Leggo and Shorthouse Reference Leggo and Shorthouse2006; Shorthouse Reference Shorthouse2010), and thus a novel approach is needed to delimit these morphologically similar species.
With advances in molecular biology, the use of molecular markers has proven essential for delimiting closely related species among Hymenoptera parasitoids (Heraty Reference Heraty2009; Santos et al. Reference Santos, Besnard and Quicke2011). The mitochondrial genome in particular serves as a good model for the study of molecular evolution and population genetics, with high rates of evolution and genome reorganisation observed in known chalcid wasp genomes (Dowton and Austin Reference Dowton and Austin1995; Oliveira et al. Reference Oliveira, Raychoudhury, Lavrov and Werren2008). A short fragment of the mitochondrial cytochrome c oxidase I (COI) gene is the core DNA barcoding animal gene and has proven successful in species identification, and distinct molecular clades or haplogroups have been used in the identification of morphologically cryptic taxa (e.g., Hebert et al. Reference Hebert, Penton, Burns, Janzen and Hallwachs2004; Smith et al. Reference Smith, Woodley, Janzen, Hallwachs and Hebert2006, Reference Smith, Wood, Janzen, Hallwachs and Hebert2007). However, controversy exists on the exclusive reliance of mitochondrial DNA in species delimitation without the inclusion of morphological or ecological datasets (e.g., Cognato Reference Cognato2006; Meier et al. Reference Meier, Shiyang, Vaidya and Ng2006). Thus, an integrative taxonomy approach is preferred using multiple independent character data sources and avoiding reliance on key characters alone in testing species hypotheses for problematic groups (Dayrat Reference Dayrat2005; Will et al. Reference Will, Mishler and Wheeler2005). Cytochrome c oxidase I has been shown to be a valuable tool in identifying cryptic taxa, in combination with morphological and ecological data, for testing host-specificity and geographical variability for Hymenoptera (e.g., Smith et al. Reference Smith, Rodriguez, Whitfield, Deans, Janzen and Hallwachs2008; Sheffield et al. Reference Sheffield, Hebert, Kevan and Packer2009; Ács et al. Reference Ács, Challis, Bihari, Blaxter, Hayward and Melika2010; Kaartinen et al. Reference Kaartinen, Stone, Hearn, Lohse and Roslin2010; Sun et al. Reference Sun, Xiao, Cook, Feng and Huang2011; Gebiola et al. 2012) including members of Eurytomidae (Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007a; Li et al. Reference Li, Zhou, Feng, Hu, Niu and Hebert2010).
The purpose this study was to use an integrative taxonomy approach to delimit eurytomids associated with galls of 14 species of Diplolepis from Canada by testing congruency of genetic variation, morphological differences, host specificity, and geographical distribution between different populations. Additionally the validity of species limits proposed by Bugbee (Reference Bugbee1967) was tested using COI sequences.
Materials and methods
Sample collection
Maturing or mature galls were collected from various sites in Canada from 1998 to 2011 (Fig. 1), either in the spring after snow melt for galls induced the previous year, or in the fall after galls had matured. Galls from the previous year were stored in jars at room temperature allowing the inhabitants to exit the galls. Galls collected in the fall were subjected to −5 °C for three to four months to break diapause. All inhabitants were either aspirated or removed with a paint brush, and then stored in 100% ethanol. The specimens used for this study were limited to those with sufficient ecological and geographical data to unambiguously identify host galls (n = 423). Eurytomids were selected from pinned specimens and bulk samples stored in 100% ethanol and identified to the species level based on dichotomous morphological keys by Bugbee (Reference Bugbee1951a, Reference Bugbee1967) in combination with host records whenever possible. Specimens that could not be confidently identified were separated into morphospecies. Localities of the eurytomids used in the study are shown in Figure 1. This map was generated using Simplemappr (Shorthouse Reference Shorthouse2012).

Fig. 1 Map of Canada indicating the sampling locations of Eurytomidae used in this study.
DNA extraction and PCR amplification of COI barcoding region
DNA extractions were performed at the Canadian Centre for DNA Barcoding (CCDB) in Guelph, Ontario, Canada using a silica-based 96-well automated extraction according to the protocol described by Ivanova et al. (Reference Ivanova, deWaard and Hebert2006, Reference Ivanova, deWaard and Hebert2007) in combination with the nondestructive voucher retrieval method described in Porco et al. (Reference Porco, Rougerie, Deharveng and Hebert2010). A series of primers were used listed in Table 1. PCR amplification and sequencing were performed according to the standard protocol used by CCDB (Ivanova and Grainger Reference Ivanova and Grainger2007a, Reference Ivanova and Grainger2007b).
Table 1 Primers used for PCR and sequencing.

Phylogenetic inference
Contigs were assembled using Sequencher version 4.5 and aligned by CLUSTALX in MEGA version 5.05 (Tamura et al. Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011) then manually checked by eye. Genetic distances were calculated in the Barcode of Life Data System (BOLD) using Kimura-2-Parameter (K2P) (Ratnasingham and Hebert Reference Ratnasingham and Hebert2007). Sequences of eurytomids with >350 base pairs were used in all analyses, with the sequences of Orthopelma mediator Thunberg (Hymenoptera: Ichneumonidae), Ormyrus rosae Ashmead (Hymenoptera: Ormyridae), and Torymus bedeguaris (Linnaeus) (Hymenoptera: Torymidae) as outgroups.
Maximum likelihood analyses were performed using the K2P distance model (Kimura Reference Kimura1980) in MEGA 5.05 and visualised as a phylogenetic tree. Branch support was assessed with 1000 bootstrap pseudoreplicates and was considered as supported when bootstrap value was >70%. Similarly, Bayesian inference using gamma-distributed rate variation across sites and a proportion of invariable sites with HKY + I + G model, as selected by JModeltest version 0.1.1 (Posada Reference Posada2008) was performed using MrBayes 3.2 (Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling and Höhna2012). Two parallel runs of four simultaneous Monte Carlo Markov chains (three heated and one cold) were run for four million generations, and trees sampled every 1000 generations. The burn-in value was set at 25% of the total sampled topologies, with the phylogeny estimated from a majority-rule consensus of the remaining trees at the threshold for clade acceptance set at 0.95. The trace files sequences and specimen information are deposited in the project Eurytomidae associated with galls of Diplolepis in Canada (project code MZEDO) on BOLD (www.boldsystems.org), and all sequences have been deposited in GenBank under accession numbers KC685087–KC685296.
Morphological study
Morphospecies were compared with the voucher specimens used for molecular study a posteriori, and sorted according to haplogroups. These specimens were chemically dried using Hexamethyldisilazane (Heraty and Hawks Reference Heraty and Hawks1998) before being point or card mounted. Scanning electronic microscopy or stereomicroscope photographs were taken using methods described by Gates and Pérez-Lachaud (Reference Gates and Pérez-Lachaud2012). The vouchers were also compared with type specimens located in the National Museum of Natural History (USNM) in Washington, DC, United States of America or the Canadian National Collection of Insects (CNCI) in Ottawa, Ontario, Canada. The DNA extracts are stored at the Biodiversity Institute of Ontario (Guelph, Ontario, Canada), while the specimens are deposited at USNM and CNCI.
Results
Cytochrome c oxidase I species delimitation
A variety of primers were used due to the difficulty in amplifying the COI region of chalcids as a result of the poly-T runs in priming region. This is also the likely cause of the low success rate, as COI sequences were obtained only from 220 of 423 specimens. Sequence lengths ranged from 223 base pairs to 632 base pairs, and show a strong A + T nucleotide bias (mean = 0.752) in comparison to C + G (mean = 0.248). Phylogenetic analyses identified eight haplogroups of eurytomids, seven of which have successfully matched identified females with male conspecifics that were morphologically unidentifiable. All haplogroups were well supported by maximum likelihood bootstrap and Bayesian posterior probabilities (Fig. 2). Both the subfamily Eurytominae (Tenuipetiolus + Eurytoma) and the genus Eurytoma were recovered as monophyletic, and Eurytoma iniquus Bugbee, Eurytoma longavena Bugbee, and Tenuipetiolus ruber Bugbee were recovered as distinct clades (Table 2). Deeply divergent lineages were revealed in Eurytoma spongiosa Bugbee, including an additional clade “E. spongiosa 2”. Eurytoma discordans Bugbee, Eurytoma acuta Bugbee, and Eurytoma calcarea Bugbee were grouped together into one genetically variable clade (Fig. 3). In addition, two rare haplogroups were found among unidentified species, Eurytoma species 1 with four specimens, and a single male specimen as Eurytoma species 2 (Fig. 2; Table 2). The intra-specific variation ranged from 0.2% to 3.8%, whereas the inter-specific divergence was 5.7–20.2% (Table 3).

Fig. 2 Phylogenetic tree for species of Eurytomidae associated with rose galls induced by Diplolepis in Canada based on cytochrome c oxidase I (COI) data. Maximum likelihood bootstrap support (first value) and Bayesian posterior probabilities (second value) are shown at each node. The scale bar represents the number of nucleotide substitutions per site. Orthopelma mediator (Hymenoptera: Ichneumonidae), Ormyrus rosae (Hymenoptera: Ormyridae), and Torymus bedeguaris (Hymenoptera: Torymidae) are used as outgroups.
Table 2 Collection locality and host information for eurytomid morphospecies and haplogroups.

Note: New records are indicated in bold.

Fig. 3 Expanded phylogenetic tree for the Eurytoma discordans complex. Codes after species identification indicates location of host galls on plant organ (LE, leaf; RO, root; ST, stem). The codes after the underscore are the collection location in Canada (AB, Alberta; BC, British Columbia; ON, Ontario). Maximum likelihood bootstrap support (first value) and Bayesian posterior probabilities (second value) are shown at each node.
Table 3 Intra-specific and inter-specific divergence for all haplogroups. Standard errors are shown in reverse of the matrix for interspecific divergence

Morphological study
In total, eight morphospecies of eurytomids were found associated with galls induced by Diplolepis from the collection sites, including five of the 10 species previously known from Canada (Noyes Reference Noyes2012). Eurytoma obtusilobae Ashmead was only observed in a single collection of galls of Diplolepis radicum (Osten Sacken) found near Kelowna, British Columbia, Canada in 1999. These specimens failed to generate sequences and were not used for this study. With the exception of T. ruber Bugbee, seven other morphospecies belong to the genus Eurytoma within the E. rosae species group, characterised by the presence of postgenal depressions and the raised adscrobal carina which forms the precoxal tooth in front of the mesocoxal cavities in lateral view (Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007a).
Key morphological characteristics traditionally used to distinguish eurytomids were found to be ambiguous, with morphological variations often correlated with size, and the absence or atrophy of key characters in the smaller specimens. Other variation included colour of the scape, forelegs and midlegs (yellow to black) and the ratio of wing vein length. New characters such as the number and arrangement of multiporous plate sensilla, and sculpturing on the petiole were useful in distinguishing male specimens, many of which were previously unidentifiable if encountered singly.
Host and geographical records
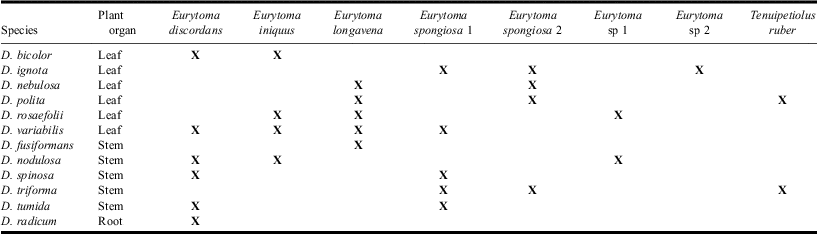
Ten new provincial records were established as a result of this study (Table 2), including a new Canadian record for E. iniquus. In addition, 18 new host associations were discovered, greatly expanding the known host records in North America. With the exception of Eurytoma species 2, which has only been observed in association with galls of Diplolepis ignota (Osten Sacken), all other haplogroups are associated with two to six different hosts (Table 4). In addition, two generations of E. longavena and E. spongiosa 2 were collected, from both spring (e.g., Diplolepis polita (Ashmead)) and fall initiated galls (e.g., Diplolepis nebulosa (Bassett)).
Table 4 Eurytomid haplogroups and associated rose galls.
Discussion
Testing species limits using COI
Accelerated rates of evolution of the chalcid mitochondrial genome have been correlated with parasitic lifestyles (Xiao et al. Reference Xiao, Jia, Murphy and Huang2011); however, testing the species limits of recently diverged lineages is difficult because the organisms often had insufficient time for the evolution of diagnostic characters or complete reproductive isolation (Xiao et al. Reference Xiao, Jia, Murphy and Huang2011; Gebiola et al. Reference Gebiola, Goméz-Zurita, Monti, Navones and Bernardo2012). Independent lines of evidence were used in the testing of species limits in these studies, thus avoiding the reliance of one particular dataset.
The COI sequences found in present study resolved closely related species of eurytomids that are difficult or impossible to distinguish morphologically. This first “screening” of morphospecies using COI drew attention to problematic clades that required further investigation (Li et al. Reference Li, Zhou, Feng, Hu, Niu and Hebert2010). An additional benefit of molecular analyses is the ability to associate sexually dimorphic eurytomids in a simple and precise way, where males were previously unknown or indistinguishable due to the lack of detailed species descriptions. The results of this study show the COI sequences are taxonomically informative in identifying eurytomid species and species boundaries defined by deep COI divergences are incongruent with morphological studies by Bugbee (Reference Bugbee1951a, Reference Bugbee1951b, Reference Bugbee1967, Reference Bugbee1973). While this study did not include additional genes, similar studies on eurytomids have shown that mitochondrial and nuclear genes corroborated each other (Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007a; Li et al. Reference Li, Zhou, Feng, Hu, Niu and Hebert2010), therefore the results of the current study based only on mitochondrial genes is likely robust when combined with ecological and host records. The species limits of three of the eight haplogroups were resolved by COI sequences although the other five haplogroups showed conflicting results with existing morphological data (Bugbee Reference Bugbee1951b, Reference Bugbee1967). The intraspecific divergence is < 2%, which is consistent with other published studies on Hymenoptera (e.g., Sheffield et al. Reference Sheffield, Hebert, Kevan and Packer2009; Li et al. Reference Li, Zhou, Feng, Hu, Niu and Hebert2010). Deep phylogenetic divergences within the COI data support the existence of cryptic genetic species in Eurytoma, consistent with previous studies of eurytomids of Palearctic gall communities (Ács et al. Reference Ács, Melika, Kalo and Kiss2002; Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007a; Gómez et al. Reference Gómez, Nieves-Aldrey, Hernández Nieves and Stone2011). The presence of cryptic species within specimens identified as E. spongiosa was expected, as the species was originally described as a morphologically variable generalist that attacked a wide variety of hosts (Bugbee Reference Bugbee1951a). A possible hypothesis for the lack of consistent morphological differences between E. spongiosa 1 and E. spongiosa 2 despite host differences could be related to the presence of Wolbachia Hertig (Rickettsiaceae), a common and widespread group of intracelluar bacteria found in the reproductive organs of arthropods that can cause cytoplasmic incompatibility (reviewed in Werren et al. Reference Werren, Baldo and Clark2008). The presence of Wolbachia is much higher within Hymenoptera in comparison to other groups of arthropods, which has been hypothesised to be the cause of host speciation (Bordenstein et al. Reference Bordenstein, O'Hara and Werren2001; Sun et al. Reference Sun, Xiao, Cook, Feng and Huang2011; Smith et al. Reference Smith, Bertrand, Crosby, Eveleigh, Fernandez-Triana and Fisher2012). As the presence of other cryptic chalcid parasitoids has been reported in association with cynipid galls (Lotfalizadeh et al. Reference Lotfalizadeh, Delvare and Rasplus2007a; Nicholls et al. Reference Nicholls, Preuss, Hayward, Melika, Csóka and Nieves-Aldrey2010), further investigation on screens for Wolbachia may help to delimit the two E. spongiosa species.
Eurytoma acuta, E. calcarea, and E. discordans were described as morphologically distinct from each other based on the colour of the scape and shape of the stigmal club and marginal vein (Bugbee Reference Bugbee1951b). Based on specimens examined in this study, these characters were shown to be extremes of a continuum rather than stable characters and are thus unreliable. The three species were also previously distinguished by their range and host, which has been expanded and now overlap as a result of additional data presented in this study. The high rate of intraspecific divergence of this clade suggests the presence of a species complex, where retention of ancestral polymorphism and hybridisation may have resulted in the failure of molecular tracing of species boundaries (Li et al. Reference Li, Zhou, Feng, Hu, Niu and Hebert2010). Hence, E. acuta and E. calcarea should be synonymised, under the more senior name E. discordans, new synonyms.
Host specificity and the evolution of eurytomids on roses
The new distribution and host records suggest that eurytomids exhibit a much wider host range than previously reported (Noyes Reference Noyes2012), as the majority of species are either oligophagous or polyphagous and were found wherever their hosts occur. Thus, using host records and range as key characteristics (Bugbee Reference Bugbee1951b, Reference Bugbee1967, Reference Bugbee1973) in species delimitation is likely prone to error. The presence of fall emergents in E. longavena and E. spongiosa 2 in both spring-initiated and late summer-initiated galls suggests that these species are bivoltine, where the first generation emerges in the spring and attacks freshly initiated galls, while the second generation develops and exits from spring galls in the late summer to attack the galls of other species of Diplolepis that are maturing at this time (Shorthouse Reference Shorthouse1973).
The radiation of Diplolepis species onto novel host plants and organs was likely in response to selection for exclusion of natural enemies (enemy-free space) such as eurytomids (Stille Reference Stille1984; Price et al. Reference Price, Fernandes and Waring1987; Stone et al. Reference Stone, Schönrogge, Atkinson, Bellido and Pujade-Villar2002). Most eurytomids are found across a wide geographical range within galls found on multiple species of wild roses; thus, their natural range likely mirrors their hosts. For instance, in cases where Diplolepis spinosa (Ashmead) shifted hosts from Rosa blanda Aiton to the domestic rose Rosa rugosa Thunberg (Shorthouse Reference Shorthouse1988), the species of eurytomid parasitoids that are normally associated with D. spinosa are also found attacking galls on the new host plant (Table 2). In a study by Nicholls et al. (Reference Nicholls, Preuss, Hayward, Melika, Csóka and Nieves-Aldrey2010), evidence was provided for parasitoids of oak galls that have tracked their hosts through space and time, showing radiation into cryptic species together with host radiations at multiple trophic levels. It is likely that eurytomids associated with rose galls also have stable, long-term co-evolutionary interactions with other species in the cynipid community, responding as a single unit to environmental perturbations (Nicholls et al. Reference Nicholls, Preuss, Hayward, Melika, Csóka and Nieves-Aldrey2010).
Several species of eurytomids examined in this study showed a close evolutionary relationship with their hosts, often only attacking hosts inducing galls on a specific plant organ (Table 4). Eurytoma longavena was observed almost exclusively in single-chambered galls such as those induced on leaves (e.g., D. polita). The only exception was galls induced by Diplolepis fusiformans (Ashmead), a small, single-chambered stem gall that is closely related to the other basal lineages of leaf-gall inducing species (Plantard et al. 1998). Likewise E. discordans was found in multi-chambered stem galls where it sometimes consumes several hosts by tunnelling from one larval chamber to another (Brooks and Shorthouse Reference Brooks and Shorthouse1997). Tenuipetiolus ruber was rarely found in galls of D. polita and Diplolepis triforma Shorthouse and Ritchie, and the intraspecific divergence of specimens identified as T. ruber (3.8%) suggests the presence of cryptic species. In addition to Diplolepis, this species has also been found in association with cynipid galls on blackberry induced by Diastrophus Hartig (Hymenoptera: Cynipidae) (Bugbee Reference Bugbee1951a). Additional specimens from other hosts to determine the species limit of T. ruber. The two unidentified species of Eurytoma and E. iniquus were collected from galls with high levels of attack by inquilines of the genus Periclistus Förster (Hymenoptera: Cynipidae) (Table 4). Eurytoma nigricoxa Provancher is the only species in Canada that has been recorded in association with Periclistus-modified galls (Bugbee Reference Bugbee1967); however, none of the three species matches E. nigricoxa upon comparison with the holotype. It is likely these three Eurytoma species are parasitoids of Periclistus; although more specimens are needed to further investigate these host relationships.
This study has established a DNA barcode reference library for eurytomids, particularly Eurytoma associated with galls of Diplolepis in Canada. This is the first phylogenetic study of Nearctic Eurytoma and suggests that many eurytomid species associated with rose galls (Bugbee Reference Bugbee1951a, Reference Bugbee1951b, Reference Bugbee1967, Reference Bugbee1973) require further investigation. Detailed studies of E. spongiosa 1, E. spongiosa 2, and the E. discordans species complex will undoubtedly aid in the identification of species. In addition, the larval forms of the eurytomids included in this study have not been described, therefore matching larvae with their corresponding adults using COI could provide valuable useful information on species delineation. Such studies of other eurytomid larvae have been morphologically informative when the adults were difficult to identify (Claridge and Askew Reference Claridge and Askew1960; Henneicke et al. Reference Henneicke, Dawah and Jervis1992; Gómez et al. Reference Gómez, Nieves-Aldrey, Hernández Nieves and Stone2011).
The presence of synonymous and cryptic species likely occurs in other eurytomid species treated by Bugbee and are in need of taxonomic revision as many morphological characters used to distinguish Nearctic eurytomids are highly variable. Issues that have impeded the identification of eurytomids associated with cynipid rose galls such as host specificity and sex association were resolved using DNA barcoding, providing new insights into the evolutionary history of this taxonomically difficult group.
Acknowledgements
The authors thank Yves Alarie, Brandy Fenwick, David Lesbarreres, and Morgan Jackson for suggestions on improving the manuscript on this and previous versions. They also thank the staff at CCDB for providing technical support with various molecular and analytical protocols. This project was supported by an Natural Sciences and Engineering Research Council Discovery Grant, a grant from the Laurentian University Research Fund, and funds from the Northern Scientific Training Program to sample galls along the shore of James Bay, awarded to JDS. Sequence analysis was carried out at the Canadian Centre for DNA Barcoding with funding from the government of Canada through Genome Canada and the Ontario Genomics Institute in support of the International Barcode of Life Project.









