Introduction
Members of the suborder Auchenorrhyncha in the order Hemiptera have needle-like sucking mouthparts and feeding exclusively on xylem/phloem plant sap. Although xylem or phloem plant sap is rich in carbohydrates, it is devoid of most essential amino acids and many vitamins (Hansen & Moran, Reference Hansen and Moran2014). Auchenorrhyncha insects are known to be associated with obligatory symbiotic bacteria, which provide essential amino acids and other nutrients for their hosts (Bressan et al., Reference Bressan, Arneodo, Simonato, Haines and Boudon-Padieu2009; Urban & Cryan, Reference Urban and Cryan2012). These endosymbiotic bacteria are vertically transmitted from mother to offspring (Ratzka et al., Reference Ratzka, Gross and Feldhaar2012) and harbored in specialized cells called bacteriocytes. The bacteriocytes typically compose a symbiotic organ called a bacteriome (Matsuura et al., Reference Matsuura, Kikuchi, Hosokawa, Koga, Meng, Kamagata, Nikoh and Fukatsu2012).
Morphological and molecular investigations have revealed that members of Auchenorrhyncha usually harbor the obligatory endosymbiont bacterium ‘Candidatus Sulcia muelleri’ (Bacteroidetes) and a partner (coprimary symbionts) belonging to the phylum Proteobacteria that are engaged in the synthesis of amino acids (Moran et al., Reference Moran, Tran and Gerardo2005; McCutcheon et al., Reference McCutcheon, McDonald and Moran2009; Urban & Cryan, Reference Urban and Cryan2012; Ishii et al., Reference Ishii, Matsuura, Kakizawa, Nikoh and Fukatsu2013; Koga et al., Reference Koga, Bennett, Cryan and Moran2013; Michalik et al., Reference Michalik, Jankowska, Kot, Gołas and Szklarzewicz2014). Urban & Cryan (Reference Urban and Cryan2012) surveyed 77 planthopper species representing 18 Fulgoroid families and detected ‘Candidatus Vidania fulgoroidea’ (Betaproteobacteria) in 40 species and the ‘Ca. Sulcia muelleri’ endosymbiont in 30 of the 40 species harboring ‘Ca. Vidania fulgoroidea’.
In addition to obligate bacterial endosymbionts, Auchenorrhyncha also harbor various facultative (secondary) bacterial endosymbionts such as Arsenophonus, Cardinium, Wolbachia, Rickettsia, Diplorickettsia and Serratia (Sacchi et al., Reference Sacchi, Genchi, Clementi, Bigliardi, Avanzati, Pajoro, Negri, Marzorati, Gonella, Alma and Daffonchio2008; Ishii et al., Reference Ishii, Matsuura, Kakizawa, Nikoh and Fukatsu2013; Hong-Xing et al., Reference Hong-Xing, Xu-Song, Ya-Jun, Jun-Ce, Qiang, Gong-Yin and Zhong-Xian2015). These bacteria are present in the cells of tissues throughout the host body and can be transmitted maternally and horizontally (Xue et al., Reference Xue, Li, Ahmed, De Barro, Ren and Qiu2012). In contrast to obligatory endosymbionts, secondary endosymbionts are not necessary for host survival (White et al., Reference White, Giorgini, Strand, Pennacchio, Minelli, Boxshall and Fusco2013); however, they do have roles ranging from neutral to pathogenic in their hosts (Oliver et al., Reference Oliver, Degnan, Burke and Moran2010). They may play a role in host fitness (Dohlen et al., Reference Dohlen, Spaulding, Shields, Havill, Rosa and Hoover2013), host resistance to natural enemies (Michalik et al., Reference Michalik, Jankowska, Kot, Gołas and Szklarzewicz2014) and host protection against environmental stresses (Hamilton & Perlman, Reference Hamilton and Perlman2013).
The date palm hopper (DPH), Ommatissus lybicus de Bergevin (Hemiptera: Tropiduchidae), is a destructive pest afflicting the date palm in the Middle East and North Africa (Hussain, Reference Hussain1963). Both nymphs and adults of this bivoltine pest cause significant damage to date palms by sucking the phloem sap. Heavy infestations of O. lybicus produce extremely large amounts of honeydew followed by the growth of sooty mold, which decreases the photosynthetic activity of the trees (Howard, Reference Howard, Howard, Moore, Giblin-Davis and Abad2001).
Excessive application of pesticides to date palm orchards to control this pest has contributed to the development of resistance of DPH populations to several conventional chemical insecticides (Ali, Reference Ali2011). To deter emergence of new resistant populations, new strategies for pest management must be developed. Interference in the endosymbiotic relationship may provide an ecofriendly method for the control of this pest (Douglas, Reference Douglas2007). Thus far, too little attention has been paid to bacterial endosymbionts in the date palm hopper. The current study investigated the diversity of bacterial endosymbionts in DPH populations. The frequency rate of bacterial endosymbionts in the studied populations was also assessed. These findings can provide insight into endosymbiotic infection of O. lybicus and open a pathway for further study of O. lybicus, its bacterial endosymbiotic interactions and possibly lead to a more ecofriendly strategy to control this pest.
Materials and methods
Collection of specimens
A total of 14 populations of DPH were collected from the major date palm growing regions of Iran and four another important date palm growing countries (Pakistan, Oman, Egypt and Tunisia) in 2015. Table 1 lists the sample collection sites and their global positioning system coordinates. All collected insects were kept in an ultra-low temperature freezer at −80°C to avoid DNA degradation until its extraction.
Table 1. Samples of Ommatissus lybicus used in this study.

DNA extraction
A single specimen was randomly selected for DNA extraction. The total DNA was extracted using the adjusted CTAB protocol from Reineke et al. (Reference Reineke, Karlovsky and Zebitz1998). Each DPH specimen was ground in liquid nitrogen and subsequently subjected to 500 µl of lysis buffer (100 mM Tris-HCl [pH 8.0], 10 mM EDTA, 2% sodium dodecyl sulfate). The homogenates were incubated in a water bath at 60°C for 1 h. After incubation, 140 µl of 5 M NaCl and 65 µl of 10% CTAB were added to the incubated homogenate and kept at 65°C for 10 min. Next, 700 µl of chloroform:isoamylalcohol (24:1) was added to each sample and they were mixed gently and placed on ice for 30 min. The mixture was centrifuged at 13,000 rpm for 20 min. After centrifugation, the supernatant was collected and 225 µl of 5 M magnesium acetate was added. The solution was mixed gently and kept on ice for 30 min, after which it was again centrifuged at 13,000 rpm for 20 min, the supernatant was discarded and 0.7 volume of cold (4°C) isopropanol was added. Samples were kept at 4°C overnight and then centrifuged for 30 min at 13,000 rpm. After centrifuging, the supernatant was removed, the pellet was washed twice with cold (4°C) 70% ethanol, dried and resuspended in 50 µl double-distilled water. The quantity and quality of the extracted DNA were determined using Thermo NanoDrop 1000 and confirmed by visualization on agarose gel (1%). The extracted DNA was stored at −20°C until use.
PCR assay and DNA sequencing
PCR assay was used to detect the bacterial endosymbionts using universal and specific primer pairs to amplify specified parts of the related bacterial gene (table 2). All PCR reactions were performed in a total volume of 30 µl containing 15 µl buffer mix, 1 µl forward primer (10 pmol µl−1) and 1 µl reverse primer (10 pmol µl−1), 1 µl DNA template and 12 µl double-distilled water.
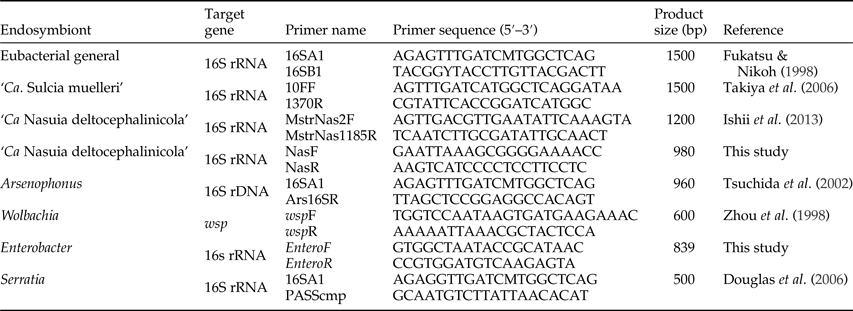
Table 2. PCR primers used to identify the bacterial endosymbionts in Ommatissus lybicus.

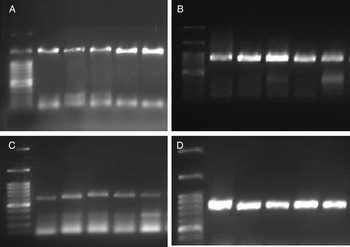
The PCR was performed in an Eppendorf thermocycler according to the PCR conditions shown in table 3. A negative control containing no DNA template was also kept with each reaction. The sample of DNA from Hishimonus phycitis Distant. (Hemiptera: Cicadellidae) was included in each PCR as a positive control for all investigated bacterial endosymbionts (Hemati et al., unpublished data). PCR products were stained with FluoroDye (Smobio; Taiwan) and subjected to electrophoresis on 1% agarose gel (fig. 1). All PCR products were directly sequenced by Macrogen Sequencing Service (South Korea). All sequences were edited using SeqMan II software (Lasergene, Version 5; DNA Star, Inc, Madison, Wisconsin, USA) and identified based on BLAST similarity searches. All sequences were deposited in the GenBank under accession numbers KX790331 to KX790338 and KY346959.

Fig. 1. Diagnostic polymerase chain reaction of the bacterial endosymbionts in Ommatissus lybicus. ‘Candidatus Sulcia muelleri‘(A: lanes 2–6), Arsenophonus (B: lanes 2–6), Wolbachia (C: lanes 2–6) and Enterobacter (D: lanes 2–6), DNA Marker (Smobio, Taiwan) (Lane 1) comprising band sizes at 3000, 1500, 1000, 900, 800, 700, 600, 500, 400, 300, 200 and 100 bp.
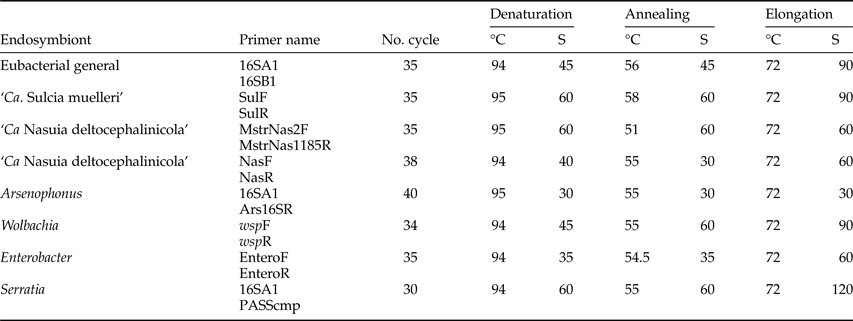
Table 3. PCR conditions to detect bacterial endosymbionts in Ommatissus lybicus.

Molecular phylogenetic analysis
Phylogenetic analysis was performed using a data set of 16S rRNA gene sequences of symbiotic bacteria downloaded from the GenBank. The phylogenetic trees of DPH bacterial endosymbionts were constructed by Bayesian analysis using the MrBayes 3.1.2 program (Ronquist & Huelsenbeck, Reference Ronquist and Huelsenbeck2003). Posterior probabilities were calculated for each node used for statistical evaluation in Bayesian analysis.
Results
Endosymbiont diversity
The PCR assay and subsequent sequencing analysis confirmed the presence of a primary endosymbiont, ‘Ca. Sulcia muelleri’ (Bacteroidetes), in all DPH populations. In addition, the presence of three secondary endosymbionts Wolbachia sp. (Alphaproteobacteria), Arsenophonus sp. (Gammaproteobacteria) and Enterobacter sp.(Gammaproteobacteria) was detected in the studied DPH populations.
By comparing the similarity of these endosymbionts with available sequences of the organism in GeneBank, different patterns of similarity were found for each endosymbiont. ‘Ca. Sulcia muelleri’ showed 96% similarity to ‘Ca. Sulcia muelleri’ endosymbiont from different species of the Fulgoridae family. The Wolbachia symbiont exhibited 100% similarity to the Wolbachia symbiont isolated from Diaphorina citri. The most similar Arsenophonus with 97% similarity has been reported as a secondary endosymbiont in Ornithomya avicularia. Two Enterobacter sequences from the Oman and Abumusa populations showed 100% similarity to the Enterobacter endosymbiont from Rhynchophorus ferrugineus and Oniticellus cinctus, respectively.
All samples were searched for the presence of ‘Candidatus Nasuia deltocephalinicola’ and Serratia, but no evidence of these endosymbionts was detected in the studied populations (table 4). Using universal primers, the bacteria Rickettsiella sp., Staphylococcus sp., Pseudomonas sp. and Erwinia sp. were detected in the DPH populations.
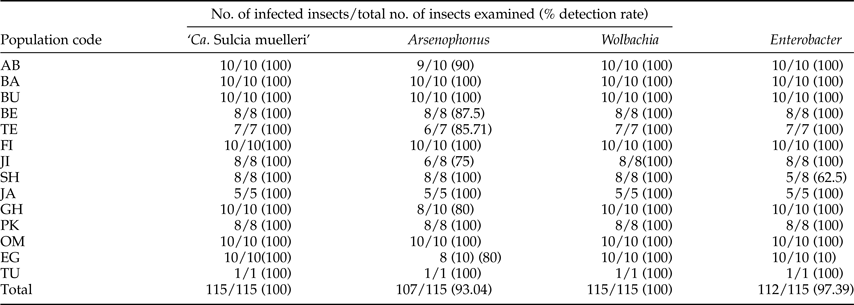
Table 4. Prevalence of bacterial endosymbionts in different Ommatissus lybicus populations.

Infection rate in DPH populations
‘Ca. Sulcia muelleri’ was detected in all DPH populations at a 100% infection rate. The average infection rate for the Arsenophonus endosymbiont was 93.04% (107 infected individuals out of 115 specimens), but its prevalence varied among populations. The highest infection rate was observed in the BA, BU, FI, SH, JA, PK, OM and TU populations. The JI population showed the lowest infection rate (six out of eight infected individuals; 75%). The secondary endosymbiont Wolbachia exhibited a 100% infection rate in all populations. The Enterobacter infection rate ranged from 62.5 to 100% in the different DPH populations (table 5).
Table 5. Infection frequencies of primary and secondary endosymbionts in different populations of Ommatissus lybicus.

Molecular phylogenetic analysis
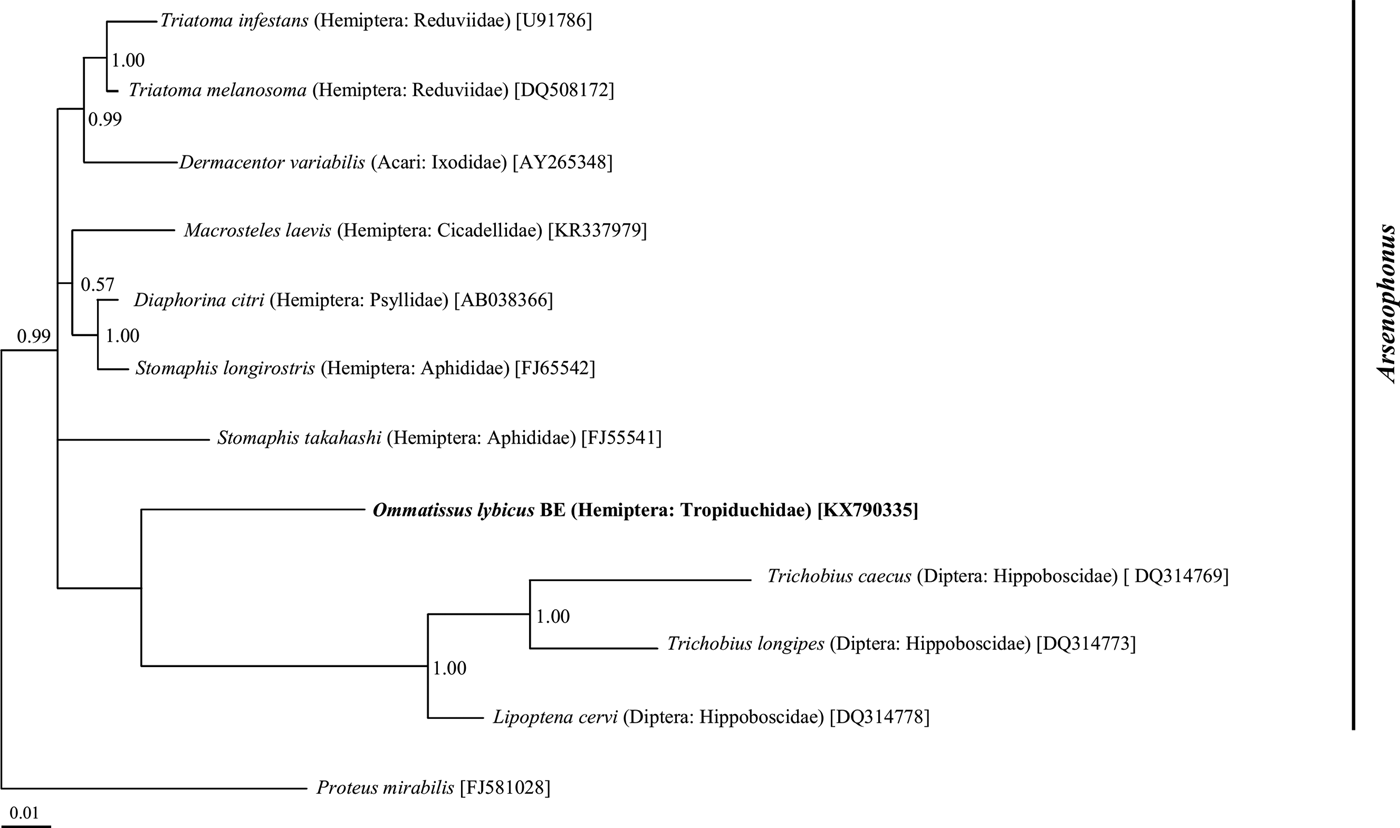
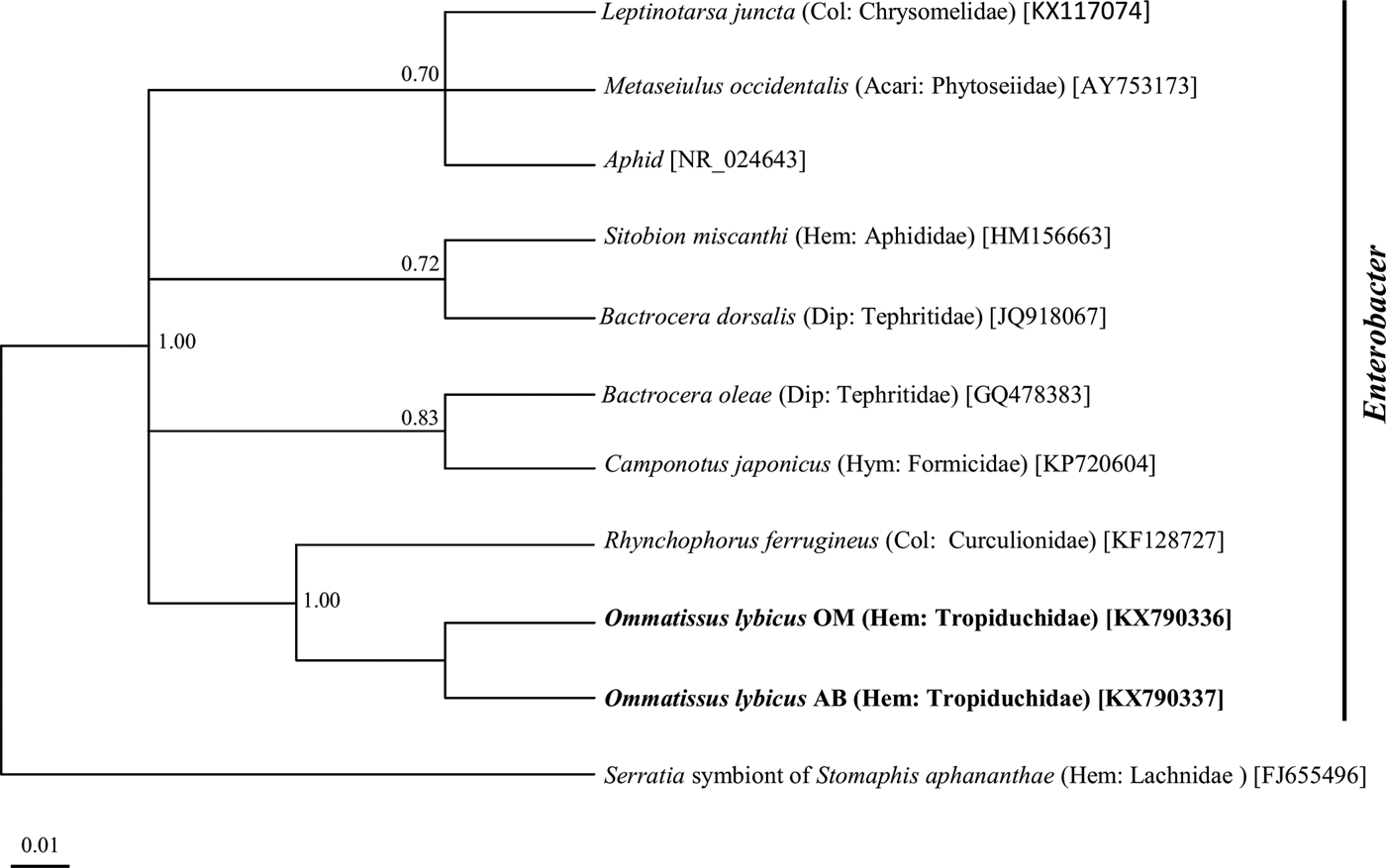
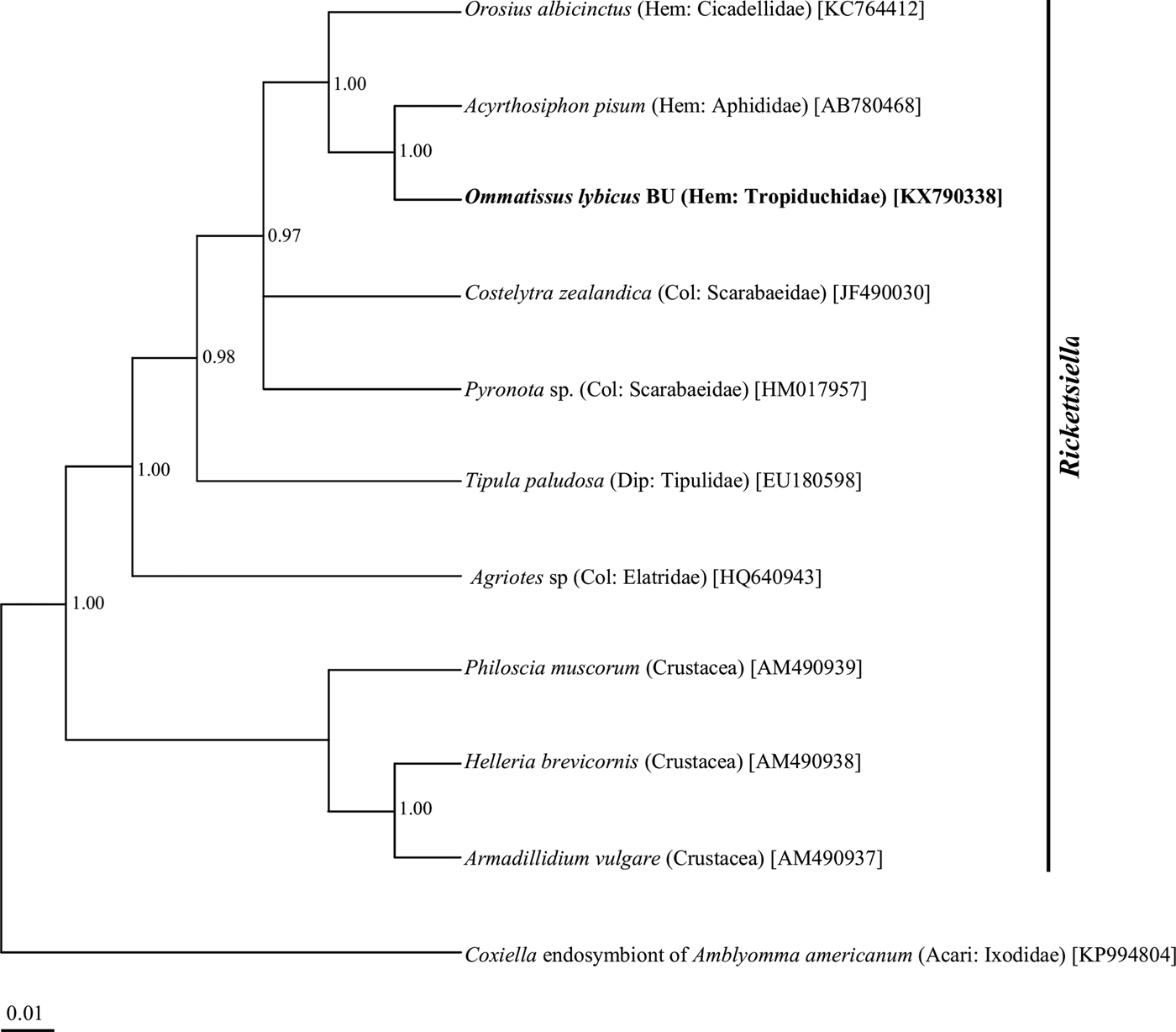
Phylogenetic analysis of the 16S rRNA gene sequences revealed a monophyletic group for ‘Ca. Sulcia muelleri’ sequenced from the DPH population in the ‘Ca. Sulcia muelleri'of the family Fulgoridae with 0.87 posterior probability values (fig. 2). The Arsenophonus symbiont of DPH was closely related to the Arsenophonus sequences from the flies of the Hippoboscidae family with 1.00 posterior probability (fig. 3). Phylogenetic analysis based on the wsp gene indicated that the DPH Wolbachia symbiont was associated with the Wolbachia sequence from Nilaparvata lugens with 0.87 posterior probability (fig. 4). The Enterobacter symbiont of DPH was closely affiliated (1.00 posterior probability) with the Enterobacter isolated from Rhynchophorus ferrugineus (Col: Curculionidae) (fig. 5). The Rickettsiella 16S rRNA sequence of DPH was closely related (1.00 posterior probability) to Rickettsiella isolated from Acyrthosiphon pisum (Hem: Aphididae) (fig. 6).
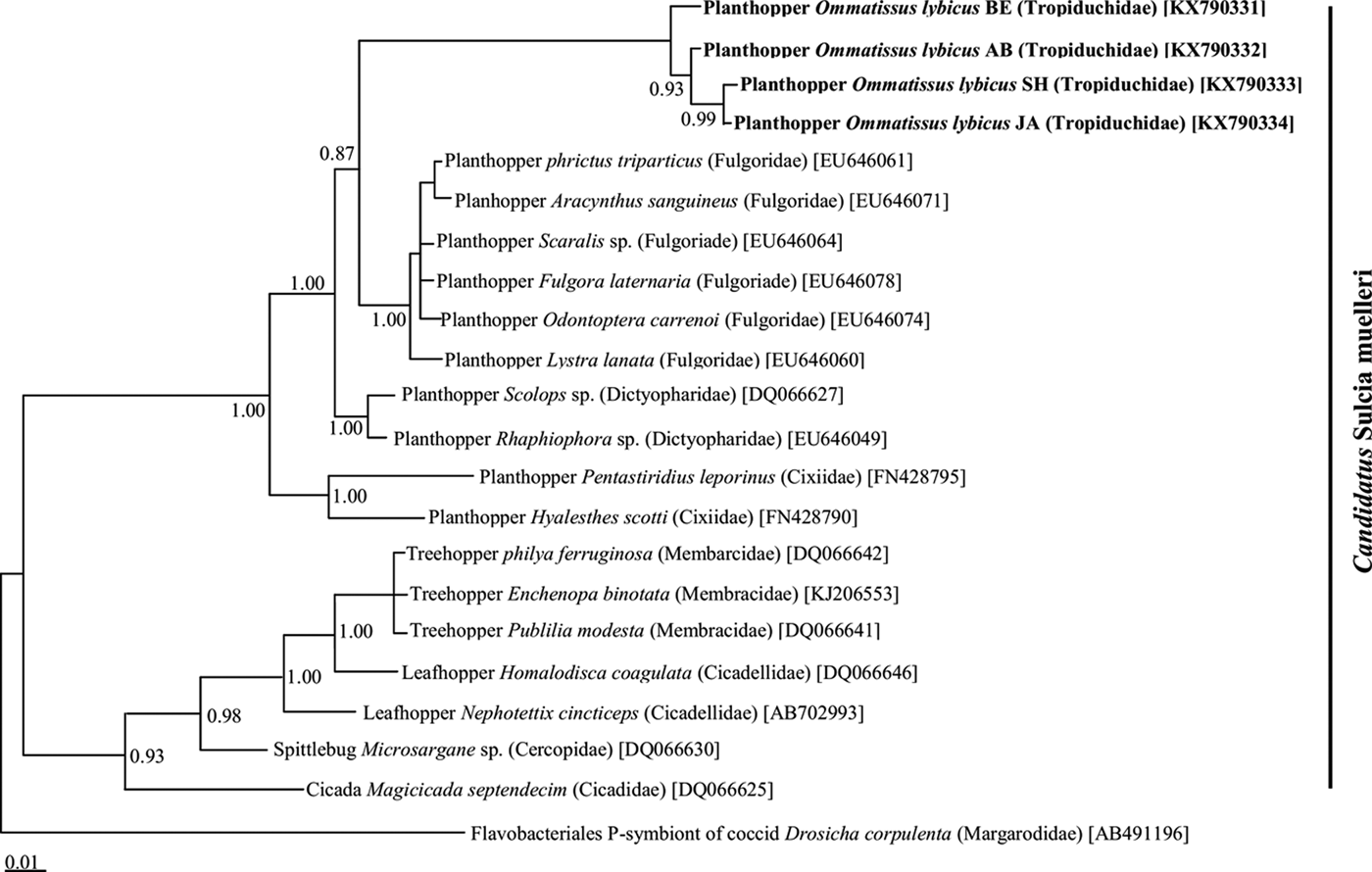
Fig. 2. Phylogenetic relationship of ‘Candidatus Sulcia muelleri’ identified from Ommatissus lybicus and other hemipteran insects on the basis of 16S rRNA gene sequences. The tree was constructed using Bayesian analysis and the numbers near nodes are posterior probabilities. The sequences obtained from O. lybicus in this study are in bold type, wherein insect species, insect family in parentheses, and GenBank accession numbers in brackets are indicated. Flavobacteriales P-symbiont of Drosicha corpulenta was used as the out group.

Fig. 3. Phylogenetic relationship of Arsenophonus symbiont identified from Ommatissus lybicus on the basis of 16S rRNA gene sequences. The tree was constructed using Bayesian analysis and the numbers near nodes are posterior probabilities. The sequences obtained from O. lybicus in this study are in bold type, wherein insect species, insect family in parentheses, and GenBank accession numbers in brackets are indicated. Sequence from Proteus mirabilis was used as out group.

Fig. 4. Phylogenetic relationship Wolbachia symbiont identified from Ommatissus lybicus on the basis of wsp gene sequences. The tree was constructed using Bayesian analysis and the numbers near nodes are posterior probabilities. The sequences obtained from O. lybicus in this study are in bold type, wherein insect species, insect family in parentheses, and GenBank accession numbers in brackets are indicated. Sequence from Anaplasma phagocytophilum was used as outgroup.
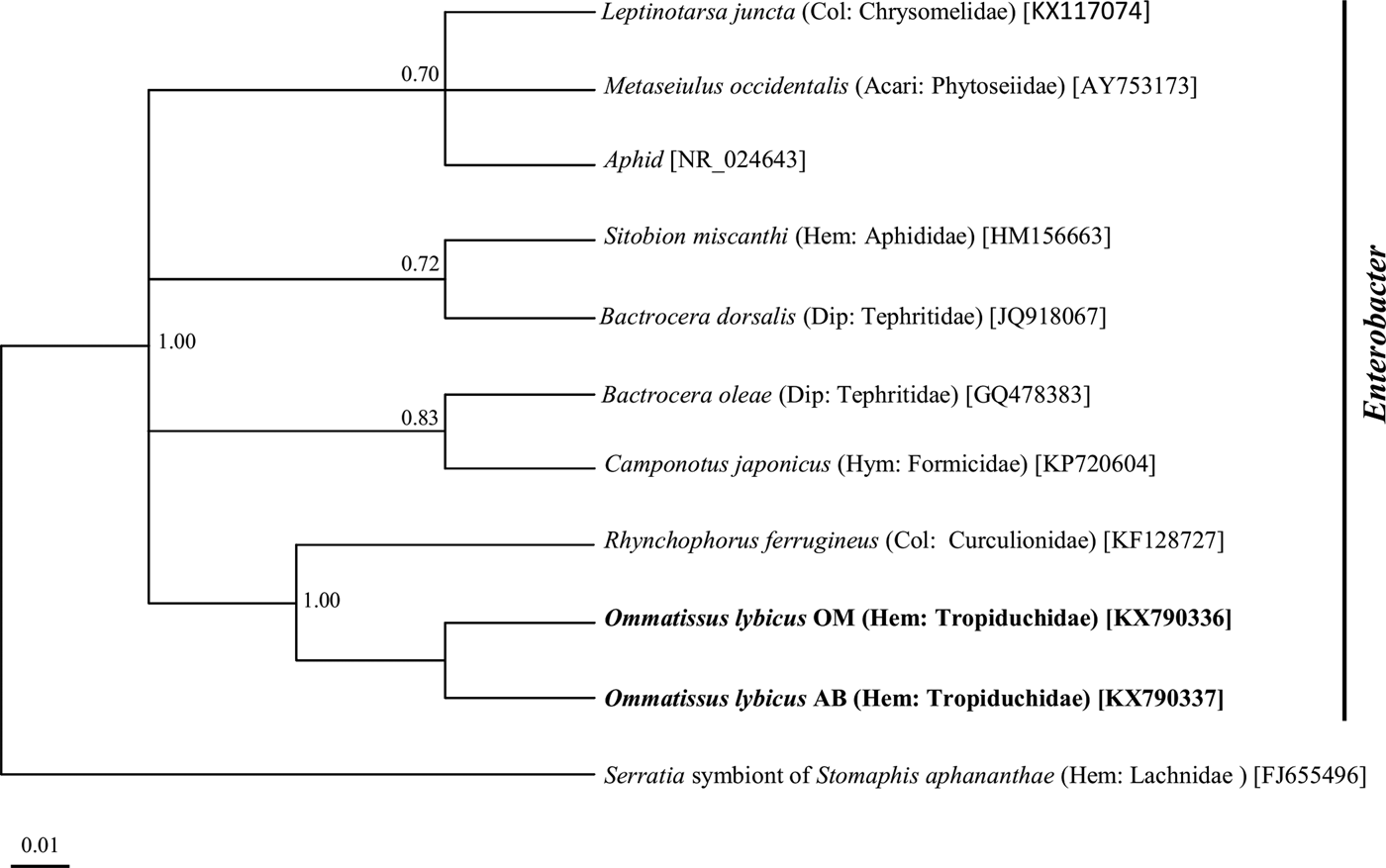
Fig. 5. Phylogenetic relationship of Enterobacter symbiont of Ommatissus lybicus on the basis of 16S rRNA gene sequences. The tree was constructed using Bayesian analysis and the numbers near nodes are posterior probabilities. The sequences obtained from O. lybicus in this study are in bold type, wherein insect species, insect family in parentheses, and GenBank accession numbers in brackets are indicated. Serratia symbiont of Stomaphis aphananthae was used as the out group.
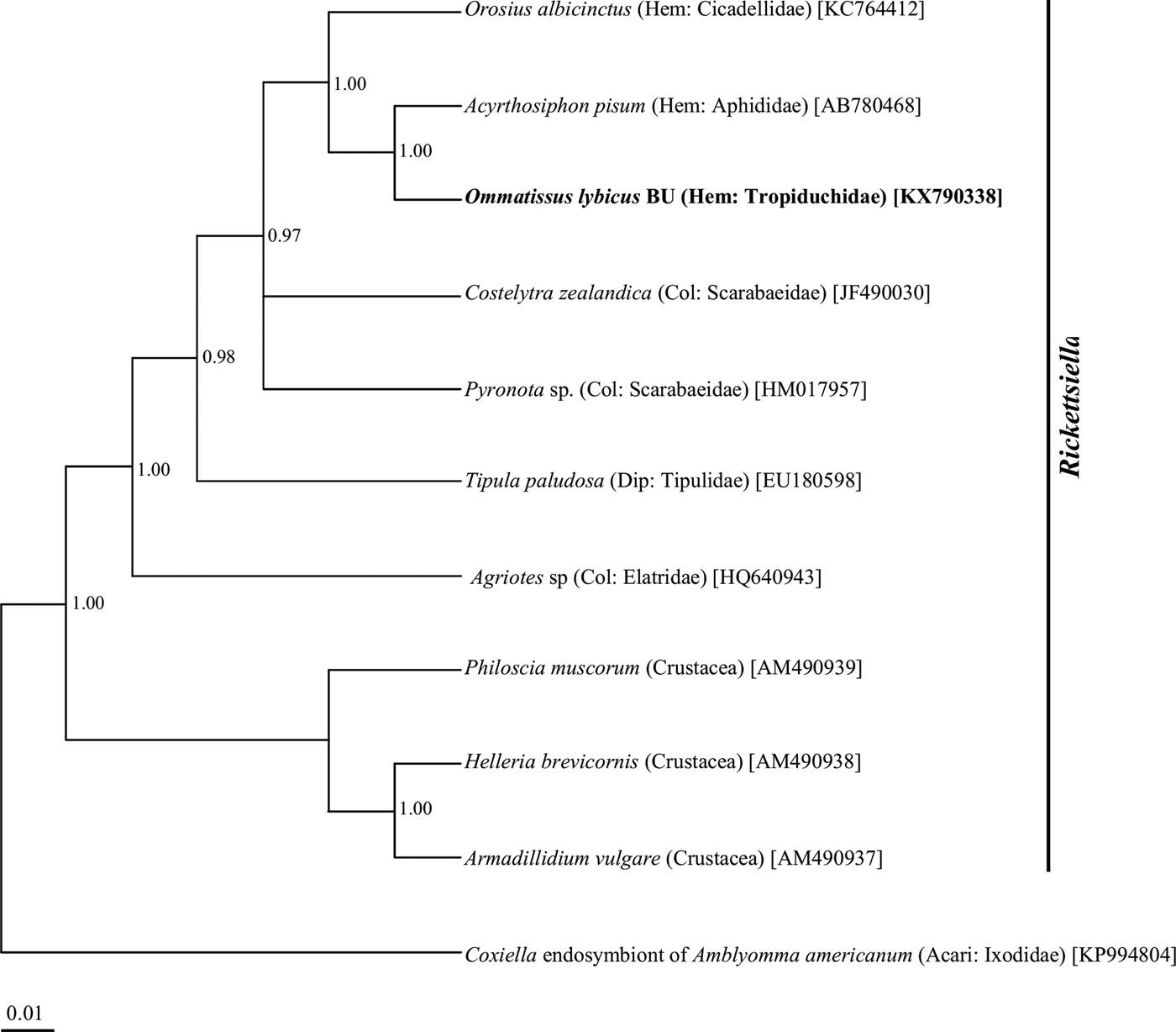
Fig. 6. Phylogenetic relationship of Rickettsiella symbiont of Ommatissus lybicus on the basis of 16S rRNA gene sequences. The tree was constructed using Bayesian analysis and the numbers near nodes are posterior probabilities. The sequences obtained from O. lybicus in this study are in bold type, wherein insect species, insect family in parentheses, and GenBank accession numbers in brackets are indicated. Coxiella endosymbiont of Amblyomma americanum was used as the out group.
Discussion
A substantial number of sap-feeding insects in the suborder Auchenorrhyncha are associated with symbiotic microorganisms (Ishii et al., Reference Ishii, Matsuura, Kakizawa, Nikoh and Fukatsu2013). Other than Cicadomorpha, little information is available about the bacterial endosymbionts of Fulgoromorpha (Urban & Cryan, Reference Urban and Cryan2012). One study revealed the presence of endosymbionts in 217 species of Fulgoroidea and showed that the a-symbiont was present in the most species (Müller, Reference Müller1940, Reference Müller1962). They hypothesized that the a-symbiont is ancient and was acquired by the common ancestor of the Fulgoroidea. Moran et al. (Reference Moran, Tran and Gerardo2005) identified ‘Ca. Sulcia muelleri’, an ancient symbiont lineage belonging to the Bacteroidetes, in 30 Auchenorrhyncha species. This matched Müller's description and illustrations of the a-symbiont.
‘Ca. Sulcia muelleri’ as an endosymbiont was also reported in Cixiidae, Delphacidae, Dictyopharidae and Fulgoridae families belonging to Fulgoroidea (Urban & Cryan, Reference Urban and Cryan2012). Several studies have demonstrated and identified the role of ‘Ca. Sulcia muelleri’ as an obligatory endosymbiont that retains genes for the synthesis of essential amino acids absent in plant sap, the food of the insects (McCutcheon & Moran, Reference McCutcheon and Moran2010). The results of the current study revealed 100% ‘Ca. Sulcia muelleri’ infection in all DPH populations, which is in line with previous findings for the Fulgoroidea superfamily. The sap-feeding behavior of the DPH lets us hypothesize that ‘Ca. Sulcia muelleri’ is similarly involved in synthesizing essential nutrients lacking in phloem sap for this species.
Recent investigations suggest that the members of Auchenorrhyncha, in addition to ‘Ca. Sulcia muelleri’, harbor other bacterial endosymbionts that complement each other in providing essential amino acids for their hosts. These include Baumannia cicadellinicola (Gammaproteobacteria) in leafhoppers (Membracoidea: Cicadellidae), Hodgkinia cicadicola (Alphaproteobacteria) in cicadas (Cicadoidea: Cicadidae) and Zinderia insecticola (Betaproteobacteria) in spittlebugs (Cercopoidea) (Koga et al., Reference Koga, Bennett, Cryan and Moran2013).
‘Ca. Nasuia deltocephalinicola’ (Betaproteobacteria) is an obligate bacterial endosymbiont, which coexists with ‘Ca. Sulcia muelleri’ in the subfamily Deltocephalinae. This bacterium produces cofactors and several essential amino acids that ‘Ca. Sulcia muelleri’ is not able to synthesize (Noda et al., Reference Noda, Watanabe, Kawai, Yukuhiro, Miyoshi, Tomizawa, Koizumi, Nikoh and Fukatsu2012; Wangkeeree et al., Reference Wangkeeree, Miller and Hanboonsong2012; Ishii et al., Reference Ishii, Matsuura, Kakizawa, Nikoh and Fukatsu2013). The current study failed to detect this endosymbiont bacterium in DPH populations. This can either be evidence of the lack of this endosymbiont in this species or that further study is required to identify its presence.
Urban & Cryan (Reference Urban and Cryan2012) detected another obligatory bacterial endosymbiont, Vidania fulgoroideae, coexisting with ‘Ca. Sulcia muelleri’ in some families belonging to Fulgoroidea. The present study did not investigate the presence of Vidania fulogoroidea in DPH populations. Because DPH belongs to this superfamily, future studies are recommended to implement such a survey.
Wolbachia, Arsenophonus and Enterobacter were detected in all 14 DPH populations. The occurrence of these endosymbionts in all DPH populations suggests significant roles for these endosymbionts on DPH population fitness and their co-evolution. Although in some populations (JA and TU), the sample size was too low to conclude 100% infection rates for the bacterial endosymbionts.
The presence of the gammaproteobacterium Arsenophonus has been evidenced in a diverse array of insects from Hemiptera, Hymenoptera and Diptera (Taylor et al., Reference Taylor, Coghlin, Floate and Perlman2011; Russell et al., Reference Russell, Funaro, Giraldo, Goldman-Huertas, Suh, Kronauer, Moreau and Pierce2012; Jousselin et al., Reference Jousselin, Coeur d'Acier, Vanlerberghe-Masutti and Duron2013; Duron et al., Reference Duron, Schneppat, Berthomieu, Goodman, Droz, Paupy, Nkoghe, Rahola and Tortosa2014). Studies have shown that this bacterium imposes significant effects on the ecology and life history of their arthropod hosts (Bressan et al., Reference Bressan, Terlizzi and Credi2012). Arsenophonus may act as a primary endosymbiont and provide essential nutrients for its host (Trowbridge et al., Reference Trowbridge, Dittmar and Whiting2006; Perotti et al., Reference Perotti, Allen, Reed and Braig2007; Nováková et al., Reference Nováková, Husník, Šochová and Hypša2015) or as a secondary endosymbiont (Chiel et al., Reference Chiel, Gottlieb, Zchori-Fein, Mozes-Daube, Katzir, Inbar and Ghanim2007). One study conferred a host protection role for Arsenophonus against parasitoid wasps (Hansen et al., Reference Hansen, Jeong, Paine and Stouthamer2007) and to promote the adaptation of the insect to specific host plants (Chiel et al., Reference Chiel, Gottlieb, Zchori-Fein, Mozes-Daube, Katzir, Inbar and Ghanim2007). Other Arsenophonus bacteria can be insect-vectored plant pathogens (Bressan, Reference Bressan2014) or parasitic agents (Gherna et al., Reference Gherna, Werren, Weisburg, Cote, Woese, Mandelco and Brenner1991).
In the current study, Arsenophonus infection was detected in all DPH populations at different frequencies. The lack of Arsenophonus in some specimens can be explained by a difference in the genetic pattern of the DPH populations, the low titer of this bacterium in the specimens or the sensitivity of the detection method. The presence of Arsenophonus endosymbiont has been reported in different populations of brown planthoppers, Nilapavata lugens (Hemiptera: Delphacidae), in recent studies (Qu et al., Reference Qu, Lou, Fan, Ye, Huang, Hu, Zhu and Zhang2013; Xu et al., Reference Xu, Zheng, Yang, Xin, Ye and Lu2014). Arsenophonus was also detected in Macrosteles laevis (Hemiptera: Cicadellidae) by Kobialka et al. (Reference Kobialka, Michalik, Walczak, Junkiert and Szklarzewicz2016).
Wolbachia has been reported from many species within Auchenorhyncha, such as Laodelphax striatellus (small brown planthopper), Sogatella furcifrea (white back planthopper) (Liu, Reference Liu2011), Nilaparvata lugens (brown planthopper) (Qu et al., Reference Qu, Lou, Fan, Ye, Huang, Hu, Zhu and Zhang2013) and Macrosteles leafhoppers (Ishii et al., Reference Ishii, Matsuura, Kakizawa, Nikoh and Fukatsu2013).
A number of effects for Wolbachia on host reproduction have been reported, including cytoplasmic incompatibility, parthenogenesis, male killing and feminization (Werren et al., Reference Werren, Baldo and Clark2008). They may increase the fitness of the insect host, provide protection against parasitization (Xue et al., Reference Xue, Li, Ahmed, De Barro, Ren and Qiu2012) and inhibit defense gene expression in plants (Barr et al., Reference Barr, Hearne, Briesacher, Clark and Davis2010).
Several studies of the genus Enterobacter have reported that this bacterium as endosymbiont from Acari (Jeyaprakash & Hoy, Reference Jeyaprakash and Hoy2010) and other insect species such as the oriental fruit fly (Bactrocera dorsalis) (Liu et al., Reference Liu, Martinez-Sañudo, Mazzon, Prabhakar, Girolami, Deng, Dai and Li2016), Bemisia tabaci (Singh et al., Reference Singh, Priya, Kumar, Rana, Ellango, Joshi, Priyadarshini, Asokan and Rajagopal2012) and rice brown planthopper (Nilaparvata lugens) (Wang et al., Reference Wang, Zhu, Lai and Fu2015).
Almost all established endosymbiotic relations of this bacterium have a beneficial function on its insect host. These include synthesis of essential nutrients (Ben-Yosef et al., Reference Ben-Yosef, Jurkevitch and Yuval2008), degradation of toxic purine compounds of the host plants (Lauzon et al., Reference Lauzon, Sjogren and Prokopy2000) and production of antiparasitic compounds like prodigiosin (Moss, Reference Moss2002). Wang et al. (Reference Wang, Chung, Peiffer, Rosa, Hoover, Zeng and Felton2016) demonstrated the suppression of plant defenses by Enterobacter bacteria in the oral secretions of the false potato beetle (Leptinotarsa juncta). All samples in the current study showed evidence of at least one secondary endosymbiotic bacteria, which is an indication of the crucial role of these organisms in the survival and fitness of DPH populations. The rapid resistance of the DPH populations to several insecticides could be closely related to these endosymbionts.
PCR assay failed to detect the Serratia endosymbiont in the studied DPH populations. This could result from the lack of this endosymbiont in these populations or the low titer of the bacterium in the tested samples. Lack of success in endosymbiont detection because of the low titer of the endosymbiont has been frequently documented (de Leon et al., Reference de Leon, Jones, Setamou and Morgan2006). Further investigation with more sensitive methods like real time PCR are needed to determine the presence/absence of Serratia.
Other groups of bacteria with less-established functions in their endosymbiotic relationships with insects were detected in some populations in this study. Members of the Rickettsiella genus have been described as facultative symbionts of diverse insects (Tsuchida et al., Reference Tsuchida, Koga, Fujiwara and Fukatsu2014). The Erwinia sp. is a common plant pathogenic bacteria and Staphylococcus sp. has been reported as an endosymbiont of some insect species (Peloquin & Greenberg, Reference Peloquin and Greenberg2003; Indiragandhi, et al., Reference Indiragandhi, Yoon, Yang, Cho, Sa and Kim2010).
Identification of major endosymbionts in insect hosts is an important step toward symbiotic control (Ricci et al., Reference Ricci, Valzano, Ulissi, Epis, Cappelli and Favia2012). The current study determined the main bacterial endosymbionts in natural populations of DPH that are assumed to effect the life history traits of their hosts. This is the first study to verify and establish the endosymbiont flora of the DPH as a key pest of the date palm with a worldwide distribution. Additional study is required to understand the role of these endosymbionts on the life history traits of the DPH and provide a pathway to establishing ecofriendly pest management through symbiotic control of this pest.
Acknowledgements
The authors like to express their special thanks to the Department of Plant Protection of Vali-e-Asr University (Rafsanjan, Iran) and the Agricultural and Natural Resources Research and Education Center of Hormozgan (Bandar Abbas, Iran) for their financial and technical support of this research project.
Disclosure
The authors declare no conflict of interest.