Introduction
The oriental latrine fly Chrysomya megacephala (Fabricius) is distributed across all continents except Antarctica (Barbosa et al., Reference Barbosa, Couri, Coelho and Avelino-Capistrano2016). The widespread spatial and temporal distribution along with often high abundances of adults suggests a wide tolerance to temperatures. This broad thermotolerance is reflected in the rather wide temperature range allowing development and survival of C. megacephala eggs and larvae from 13 to 35°C (Alonso et al., Reference Alonso, Souza, Linhares and Thyssen2015; Gruner et al., Reference Gruner, Slone, Capinera and Turco2017). Due to the wide thermal scope, as well as other traits like ease of rearing, C. megacephala has been proposed to be used as a beneficial insect for pollination of high-value plant crops (Wang et al., Reference Wang, Zhao and Lei2005; Ramírez and Davenport, Reference Ramírez and Davenport2016; Luo, Reference Luo2018). For example, flies have been observed to visit and pollinate fruit crops during adverse weather when honeybees were absent (Wang et al., Reference Wang, Ma, Cui, Wang, Lu and Qin2011; Ellis et al., Reference Ellis, Feltham, Park, Hanley and Goulson2017). While C. megacephala does seem to possess desirable thermal traits, low temperature does constitute a limit for locomotion and survival of C. megacephala (Sontigun et al., Reference Sontigun, Sukontason, Klong-Klaew, Sanit, Samerjai, Somboon, Thanapornpoonpong, Amendt and Sukontason2018). The increase of C. megacephala cold hardiness/tolerance by experimental operations may improve the potential of this species as a pollinator of crops under unfavorable weather conditions and thus facilitate sustainable and efficient agricultural production.
Rapid cold hardening (RCH) and long-term cold acclimation (LCA) are potential treatments for achieving improved locomotion, survival, and reproduction in mass-reared beneficial insects when released into low-temperature environments (Kristensen et al., Reference Kristensen, Hoffmann, Overgaard, Sorensen, Hallas and Loeschcke2007; Boersma et al., Reference Boersma, Boardman, Gilbert and Terblanche2019). Physiological adaptations to cold include RCH and LCA, which are induced by exposure to appropriate non-lethal low temperatures, and can improve both critical thermal limits and performance at low temperatures (Teets and Denlinger, Reference Teets and Denlinger2013; Shearer et al., Reference Shearer, West, Walton, Brown, Svetec and Chiu2016; Gerken et al., Reference Gerken, Eller-Smith and Morgan2018; Izadi et al., Reference Izadi, Mohammadzadeh and Mehrabian2019). For example, acclimation at non-lethal low temperature for either 7 days during adulthood or the whole life stage of Drosophila suzukii adults decreased the chill coma recovery time and increased survival to acute cold stress (Enriquez et al., Reference Enriquez, Renault, Charrier and Colinet2018). Meanwhile, cold acclimation has been applied in pest management and beneficial insect utilization. For example, the increase of Thaumatotibia leucotreta flight performance by cold acclimation was useful for enhancing the efficacy of the sterile insect technique under cooler environmental conditions (Boersma et al., Reference Boersma, Boardman, Gilbert and Terblanche2019), and cold acclimation is advantageous for predation of Gaeolaelaps aculeifer before inoculative release in early spring (Jensen et al., Reference Jensen, Sorensen and Holmstrup2019). In C. megacephala, cold acclimation could lead to improve pollination efficiency, particularly at low temperature. While LCA has a strong impact on low-temperature performance in most insects (Teets and Denlinger, Reference Teets and Denlinger2013), this treatment imposes logistical challenges for mass rearing pollinators due to the longer development times at mildly low temperatures. Alternatively, RCH can improve cold tolerance, and potentially performance, at low temperature with treatments of much shorter durations (Shintani and Ishikawa, Reference Shintani and Ishikawa2007). If RCH substantially increases cold performance of mass-reared pollinators, this might make conditioning insects intended for release during cool periods more logistically feasibly and efficient.
High-throughput metabolomics and transcriptomics have revealed many mechanisms involved in insect cold acclimation responses. Accumulation of cryoprotectants (polyols, free amino acid, etc.) and up-regulation of heat shock proteins are important mechanisms mediating improved cold tolerance in response to cold acclimation (Rinehart et al., Reference Rinehart, Li, Yocum, Robich, Hayward and Denlinger2007; Colinet et al., Reference Colinet, Lee and Hoffmann2010; Schou et al., Reference Schou, Kristensen, Pedersen, Karlsson, Loeschcke and Malmendal2016; Enriquez et al., Reference Enriquez, Renault, Charrier and Colinet2018). In addition, cold-acclimated Drosophila melanogaster had low activity of Na+/K+-ATPase, which contributed to the maintenance of low hemolymph [Na+], and played a vital role for cold tolerance (MacMillan et al., Reference MacMillan, Ferguson, Nicolai, Donini, Staples and Sinclair2014). In Gryllus pennsylvanicus, ion-regulatory tissues had reduced chilling injury after cold acclimation (Des Marteaux et al., Reference Des Marteaux, McKinnon, Udaka, Toxopeus and Sinclair2017). As for RCH, the increased survival to acute cold stress seems to be mediated by mechanisms shared with those of cold acclimation (Izadi et al., Reference Izadi, Mohammadzadeh and Mehrabian2019), e.g. ion transport mechanisms (Armstrong et al., Reference Armstrong, Rodríguez and Meldrum2012; MacMillan et al., Reference MacMillan, Ferguson, Nicolai, Donini, Staples and Sinclair2014; Coello Alvarado et al., Reference Coello Alvarado, MacMillan and Sinclair2015; Overgaard and MacMillan, Reference Overgaard and MacMillan2017) and accumulation of cryoprotectants (Storey et al., Reference Storey, Baust and Storey1981; Park and Kim, Reference Park and Kim2014). However, there also seem to be huge differences between responses to RCH and LCA (Gerken et al., Reference Gerken, Eller, Hahn and Morgan2015). Some genes and pathways related to signal transduction are activated by RCH, e.g. calcium signaling mediated cold sensing (Teets et al., Reference Teets, Yi, Lee and Denlinger2013) and p38 MAPK played an important role in signal transduction triggering RCH in the flesh fly Sarcophaga crassipalpis (Fujiwara and Denlinger, Reference Fujiwara and Denlinger2007). Although the mechanisms of RCH and LCA have been compared in numerous studies across various insect systems, few studies directly compare transcriptomic responses of both RCH and LCA.
Here we investigated the impact of RCH and LCA on cold tolerance of C. megacephala, in order to evaluate the potential for improving low-temperature performance and compare the underlying molecular mechanisms of those two processes. We carried out transcriptomic analysis of RCH and LCA responses by Illumina sequencing technology. The present findings suggested that different sets of physiological pathways are activated by RCH vs. LCA and there were more transcripts and pathways associated with LCA than RCH. RCH and LCA both offer promise for increasing cold tolerance of C. megacephala and other beneficial insects to extend their use at lower temperatures in the near future.
Materials and methods
Insect rearing and maintenance
Eggs of C. megacephala were collected in Huazhong Agricultural University (HZAU), Wuhan, P. R. China (30°4′N and 114°3′ E), using chicken meat as baits. These eggs and other stage individuals were maintained in the same artificial climate incubator (Ruihua, China) at 25 ± 1°C with 60 ± 5% RH and LD cycle of 12 h L/12 h D. Larvae were fed on artificial diets (wheat bran, fish power, chicken liver and water, w:w:w:w = 7:2:1:20) until pupation in a plastic container. Pupae were sifted from the artificial diet residue and placed in a plastic bowl which was put in the same incubator. Adults were reared in mesh cages (30 cm × 30 cm × 50 cm) and supplied with ad libitum access to sugar powder, chicken meat, and water.
Discriminating temperature
Adults were maintained at 25 ± 1°C until low-temperature assay. Survival of adults was assessed under different temperatures (−9 to −2°C) for 2 h. Groups of 20–30 adults (both males and females together) were placed in 50 ml plastic containers. The containers were submerged in ethanol-water mixture (v:v = 7:3) in a low temperature bath (DC-3006, SCIENTZ, China). After cold treatment, adults were allowed to recovery under 25°C for 24 h. Survivals of female and male were assessed separately by the ability of flies to right themselves after being turned on their back 24 h after the initial cold shock. Based on survival, a discriminating low temperature (survival closed to 20%) for C. megacephala adults was chosen (−6°C) for the following experiments.
RCH and LCA pre-treatments and cold tolerance
Adults 2 days post-eclosion were divided into two groups: RCH and LCA. Adults were exposed to either direct cold shock (direct transfer from 25 to −6°C for 2 h) or RCH (transfer from 25°C to ice-water mixture for 1, 2, 4, or 8 h and then immediately transferred to −6°C for 2 h). Cold tolerance was assessed by survival as described above. For LCA, adults were divided into the following groups: (1) control or no acclimation (LC), maintained at 25°C; (2) fluctuating thermal regime (FTR) where temperature oscillated daily: the climate incubator was set at 15/25°C (12/12 h), but there was a short lagtime for the temperature change; (3) the stable cold regime (SCR) which were maintained at 15°C. After 7 days, cold tolerance (measured as survival as described above) of adults from each group was assayed after exposure to −6°C for 2 h. Each treatment was replicated four times and for each replicate, 15–25 individuals were used. Another group of adults that had spent 7 days in each treatment were used to measure chill coma recovery time as an additional metric of cold tolerance. Chill coma was induced by placing groups of adults into an ice-water mixture for 2, 4, or 6 h, followed by recovery at 25°C. Recovery time for each fly was defined as the number of seconds from the time flies were removed from the ice-water slurry until a fly could right itself and stand up.
RNA isolation and cDNA library construction
For RCH, control females (RCF) and males (RCM) were maintained at 25°C; R30F and R30M referred to the female and male samples collected during early stage of RCH (at ice-water mixture for 30 min); and RF and RM referred to the female and male samples kept at ice-water mixture for 2 h followed by recovery at 25°C for 30 min (fig. 1a). For LCA, only males were sampled for transcriptomic analysis, as disruption of females oviposition upon cold stress might affect transcriptomic results. The samples of FTR males were collected at the end of 15°C and temperature will shift to 25°C gradually in the next short time (fig. 1b). LC, FTR, and SCR male samples were collected after 7 days and then frozen rapidly in liquid nitrogen, followed by storage at −80°C until the RNA extraction. Each treatment was replicated three times and for each replication, a pool of ten individuals was used.

Figure 1. Temperature regimes and RNA collection time points. (a) Chrysomya megacephala adults were transferred from 25 to 0°C for 2 h, followed by recovery at 25°C. Samples contain individuals during early stage (transfer from 25 to 0°C for 30 min) and recovery stage of RCH (transfer from 0 to 25°C for 30 min) and control (kept at 25°C). The black solid line represents temperature fluctuation during RCH and recovery period, and these red spots represent sampling time point of different treatments. R30F, females during the early stage of RCH; R30M, males during the early stage of RCH; RF, females during the recovery stage of RCH; RM, males during the recovery stage of RCH; RCF, control females; RCM, control males. (b) Adults were reared in a fluctuating thermal regime (FTR): 15/25°C (12/12 h), a stable cold regime (SCR) (15°C), or a stable warm control (LC) (25°C) for 7 days.
Total RNA was extracted by Trizol reagent (Invitrogen, Carlsbad, CA, USA) following manufacturer's instructions and genomic DNA was removed using DNase I (Takara, Dalian, China). RNA quality was determined with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and gel electrophoresis, and quantified using a NanoDrop-2000 (Thermo Scientific, Wilmington, DE, USA). Only high-quality RNA (2.3 > OD260/280 > 1.9, OD260/230 > 1.5, RIN > 7, 28S : 18S > 1.0) was used to construct sequencing libraries. Reverse transcription, library construction, and sequencing were performed according to the manufacturer's instructions (Illumina, San Diego, CA, USA). The C. megacephala RNA-seq transcriptome libraries were generated using the Illumina TruSeqTM RNA sample preparation Kit from Illumina® (NEB, USA) following manufacturer's recommendations. After quality inspection of cDNA samples, library preparations were sequenced on an Illumina Hiseq 2500 system (Illumina) and paired-end reads (2 × 150 bp) were generated in FASTQ format.
De novo transcriptome assembly and gene annotation
Raw reads were filtered to remove low-quality reads by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle). Reads with an adapter, poly-N, or those shorter than 20 bp were removed from the data set. A pool of reads was formed by merging all 27 samples of sequencing data. De novo transcriptome assembly used Trinity (Version v2.8.5, http://trinityrnaseq.sourceforge.net/) (Grabherr et al., Reference Grabherr, Haas, Yassour, Levin, Thompson, Amit, Adiconis, Fan, Raychowdhury, Zeng, Chen, Mauceli, Hacohen, Gnirke, Rhind, di Palma, Birren, Nusbaum, Lindblad-Toh, Friedman and Regev2011). CD-HIT (Version v4.5.7, http://weizhongli-lab.org/cd-hit/) was used to exclude redundant sequences and combine sequences with more than 90% similarity and an overlapping length longer than 35 bp. The completeness assessment was performed by TransRate (Version v1.0.3, http://hibberdlab.com/transrate/) and BUSCO (Version 3.0.2, http://busco.ezlab.org). Contigs longer than 200 bases were used for subsequent analysis. Gene function was assigned based on the following annotation databases: NR (NCBI non-redundant protein sequences) (Version 2019.6.26), Swiss-Prot (a manually annotated and reviewed protein sequence database) (Version 2019.7.1), PFAM (protein family) (Version v32.0), COG (Clusters of Orthologous Groups of proteins) (eggNOG database Version 5.0), and KEGG (Kyoto Encyclopedia of Genes and Genomes Orthology) (Version 2017.08) using BLASTX (E-value <0.00001). Blast2GO (http://www.blast2go.com/b2ghome) was used to obtain gene ontology (GO) annotations of unigenes (unique assembled transcripts by redundancy removal, which represents each unigene is a unique sequence of one gene) to describe biological processes, molecular functions, and cellular components.
Differentially expressed genes (DEG) profiling
Through mixing and splicing mRNA sequences of all 27 samples of C. megacephala, we obtained a transcriptome database for reference sequences. The clean reads for each sample were matched with the reference sequences by RSEM software (Version 1.3.1) with default values (Li and Dewey, Reference Li and Dewey2011). The FPKM (expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) method was used to calculate expression counts of unigenes and multiples of differential expression levels among samples for a gene. We used the DESeq (Version 1.24.0) to screen DEGs among two treatments. |log2 (fold change)| ≥ 1 and adjusted P-value <0.05 were used as the parameters. In addition, KEGG database analyses were performed to identify metabolic pathways in which the DEGs were significantly enriched at a Bonferroni-corrected P-value <0.05 compared with the whole transcriptome background. KEGG pathway enrichments were carried out using Goatools (Version 0.6.5, https://github.com/tanghaibao/Goatools). Heatmaps were constructed in the R environment (R 3.4.2, R package: pheatmap). DEGs were divided into different clusters by expression trend using Genesis based on the K-means method (Sturn et al., Reference Sturn, Quackenbush and Trajanoski2002). Pathway enrichment analysis of up-regulated genes was carried out based on the KEGG database. Related custom scripts of bioinformatics analysis have been stored in Github (https://github.com/qixuewei1/Transcriptome-analysis-codes).
IPath2.0 (http://pathways.embl.de) was used for the visualization of metabolic pathways. KEGG Orthologs (KO) IDs of up- and down-regulated genes were submitted to this website.
Quantitative real-time PCR (qPCR) validation
We validated the estimates of gene expression levels in different treatments for seven genes using qPCR, i.e. Death-associated inhibitor of apoptosis 1-like (DIA1), Filaggrin-2 (F2X2), Growth arrest and DNA damage-inducible protein GADD45 alpha (GADD45), HSP40, HSP67, Spectrin alpha chain (SACX1) and Dihydropyrimidinase-like (DPLX1). These genes were randomly selected from DEGs shared by SCR and RF compared with their respective controls. Briefly, 2 μg of total RNA (prepared above) was used for reverse transcription using PrimeScript RT reagent Kit (Takara). qPCR were run on three biological replicates and four technical replicates for each treatment. Premier 5.0 (Premier Biosoft International, Palo Alto, CA, USA) was used to design primers (table S1). The stability of eight candidate reference genes (Wang et al., Reference Wang, Xiong, Wang, Lei and Zhu2015a) was assessed by exposing adults to different temperatures (0°C for 2 h; 15, 20, and 25°C for 7 d). Normfinder, and geNorm showed that β-tub (F: CCATTTCATCCATACCCTCACC; R: GCTTGAAAATGTCTGCCACC) was the most stable transcript (fig. S1), so β-tub was chosen as an internal reference gene for this study. The PCR reaction protocol was as follows: 95°C for 30 s, 40 cycles at 95°C for 5 s, 55°C for 30 s, and 72°C for 31 s. All data were imported into Microsoft-Excel (Microsoft Excel 2016, USA) and analyzed via the 2−ΔΔCT method (Livak and Schmittgen, Reference Livak and Schmittgen2001). The correspondence between the gene expression data obtained by transcriptome and qPCR were investigated using a linear model on log-transformed data.
Statistical analysis
The data on survival after exposure to −6°C after RCH with increasing acclimation time were investigated using a Gauss curve fit model containing survival as a response variable and acclimation time as an explanatory variable, with the Levenberg–Marquardt algorithm used to obtain fit non-linear model parameters. The relationship between chill coma recovery time and the duration of exposure to chill coma-inducing temperatures was investigated using a linear model containing recovery time as a response variable and exposure time as an explanatory variable. All data were tested for homogeneity of variances using Levene's tests. One-way analysis of variance (ANOVA) was performed to analyze data on survival after RCH and the recovery time after LCA using SPSS 16.0 (SPSS Inc., Chicago, IL, USA). Two-way ANOVA was performed for tests of sex and acclimation type on survival after LCA. Differences among measured parameters were considered significant when P-values were <0.05 after Tukey's Honestly Significant Difference (HSD) correction for multiple comparisons. The gene relative expression of RCH and LCA-treated individuals relative to controls was determined by two-tailed Student's t-test at the significance levels at *P < 0.05, **P < 0.01, ***P < 0.001. All results were expressed as means with standard errors (SE).
Results
Discriminating temperature of C. megacephala adults
Survival of C. megacephala adults after 2 h exposure was significantly different among different exposure temperatures (female: df = 6, F = 129.1, P < 0.001; male: df = 6, F = 113.1, P < 0.001) (fig. 2a, b). Survival of female and male was 22.9 and 26.4%, respectively, by exposure to −6°C for 2 h, so −6°C was recognized as discriminating temperature for adults and could be used for exposure temperature to detect survival in subsequent experiments.
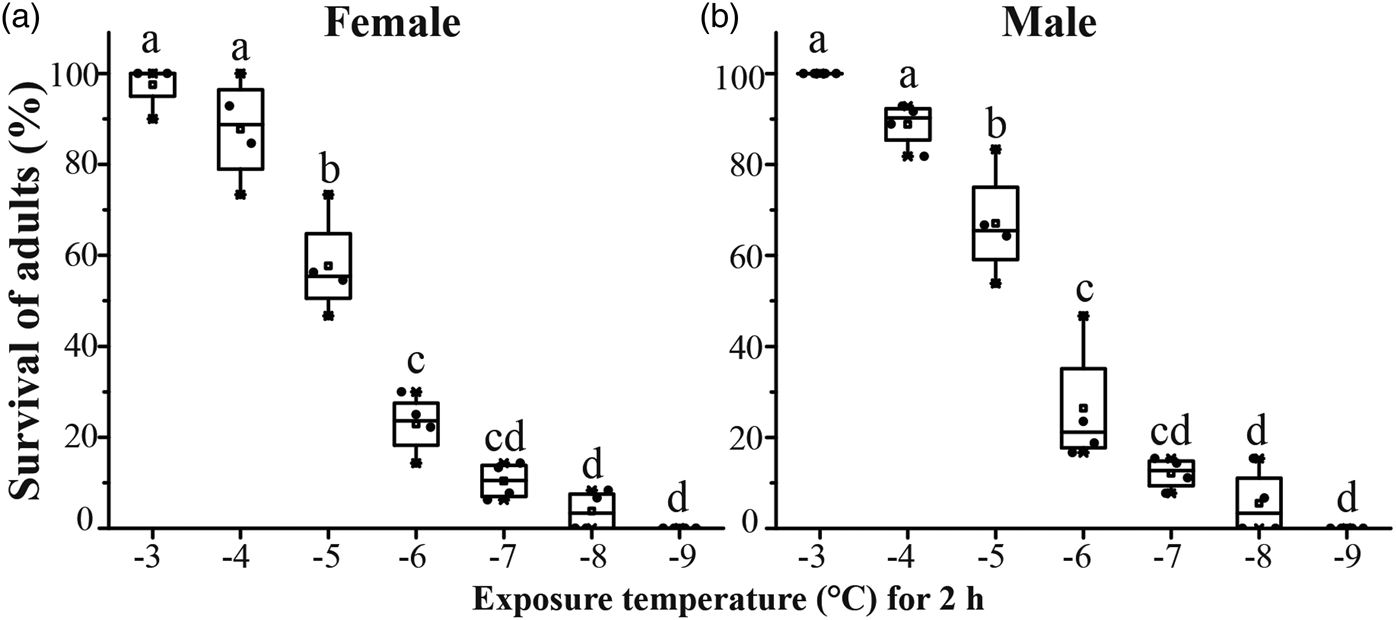
Figure 2. Survival of Chrysomya megacephala female (a) and male (b) exposed to different low temperatures. Adults were exposed to different temperatures (−9 to −2°C) for 2 h. After cold treatment, adults were allowed to recovery at 25°C for 24 h. Survivals of female and male were assessed by the ability of flies to right themselves after being turned on their back. Distinct letters denote groups that are significantly different from each other after Tukey's HSD correction for multiple comparisons.
Cold survival of C. megacephala after RCH
Both sexes of C. megacephala showed a robust RCH response between 1 and 4 h of pre-exposure to 0°C before cold shock at −6°C that was highest at 2 h (fig. 3a, females, df = 5, F = 25.23, P < 0.001; fig. 3b, males, df = 5, F = 23.85, P < 0.001). However, pre-exposure periods of 6 and 8 h appeared to themselves to be stressful because survival after cold shock dwindled to the point that survival was equal to or lower than non-exposed controls.

Figure 3. Cold tolerance of Chrysomya megacephala adults after RCH or LCA. Survival of C. megacephala adults (a: female; b: male) after RCH (0°C for 1, 2, 4, 6, 8 h, respectively) prior to cold shock at −6°C for 2 h, control adults were placed directly at −6°C directly. Distinct letters denote groups that are significantly different from each other after Tukey's HSD correction for multiple comparisons. Grey lines show a Gauss curve fit after different RCH times (female: reduced χ 2 = 1.573, R 2 = 0.977; male: reduced χ 2 = 3.363, R 2 = 0.948). LCA contains FTR (fluctuating thermal regime: 15/25°C for 7 days) and SCR (stable cold regime: reared in 15°C for 7 days); the adult reared in 25°C for 7 days as control. Recovery time was detected at 25°C after exposure to 0°C for 2h, 4h, or 6 h (c: female; d: male). Lines show recovery time increased with exposure time. The survival of C. megacephala (e) after cold shock of −6°C for 2 h was analyzed by two-way ANOVA with sex and acclimation type as mix factor. Groups treated within the same exposure time with different letters are significantly different (P < 0.05; Tukey HSD).
Recovery time and cold survival of C. megacephala after LCA
In females, both cold-acclimation treatments (FTR and SCR) decreased recovery time compared to flies reared at constant warm temperature for all three of our chill-coma durations (P < 0.05) (fig. 3c). In males, both cold-acclimation treatments (FTR and SCR) decreased recovery time compared to flies reared at constant warm temperature for 4 and 6 h-chill coma durations (P < 0.05) while there was no significant difference for 2 h between SCR and control (P > 0.05) (fig. 3d). Furthermore, acclimation treatments (FTR and SCR) also substantially increased survival after a cold shock at −6°C for 2 h compared to flies reared at constant warm temperature in both sexes (df = 2, F = 18.4, P < 0.001). Moreover, these was no significant effect of sex and of the interaction between sex and acclimation type on survival after LCA (sex: df = 1, F = 1.1, P = 0.319; interaction: df = 2, F = 0.5, P = 0.617) (fig. 3e).
Illumina sequencing and assembly
The transcriptome of all samples combined contained an average of 54,633,149 raw reads. After removing low-quality sequences, 54,260,866 clean reads remained. The Q20 quality score of every sample was >97.7% (table S2). De novo transcriptome assembly with Trinity using reads from all samples generated 58,822 transcripts and 42,692 unigenes. The average length of the unigenes was 1075 bp (ranging from 201 to 31,666 bp) with an N50 = 1764 bp. Detailed sequencing and assembly results are provided in tables S2 and S3. Of the unigenes, 72% had a length between 200 and 1000 bp (table S4). Meanwhile, 12,160 unigenes had a length more than 1000 bp and 5163 unigenes had a length more than 2000 bp (table S4).
Annotation of unigenes
In total, 24,397 (57.2%) unigenes were found in at least one public database, such as the NCBI Nr (non-redundant protein sequences), Swiss-Prot, PFAM, COG, GO, and KEGG (KEGG Orthology) (table S5). Based on the Nr annotation, about 72.3% unigenes had strong homology with e-values <10−30 (fig. S2A), and 87.2% showed a homology above 60% with known mRNA sequences (fig. S2B). Among annotated unigenes, the top matched species was Lucilia cuprina with 13,174 unigenes (61.6%) (fig. S2C). Lucilia cuprina is also a blowfly that is an important pollinator in some contexts (Munawar et al., Reference Munawar, Raja, Sarwar and Niaz2011; Brodie et al., Reference Brodie, Smith, Lawrence and Gries2015).
Functional classification of unigenes by GO and KEGG
A total of 15,943 unigenes (37.3%) were assigned GO annotations for functional classification (fig. S3A). Overall, 13,579 (31.8%) of unigenes were assigned KEGG annotations and divided into six main categories: metabolism environmental information processing, genetic information processing, cellular processes, organismal systems, and human diseases. The top three subcategories were signal transduction (1542 genes); translation (1530 genes); and folding, sorting, and degradation (961 genes) (fig. S3B).
DEGs of C. megacephala adults during RCH
Among DEGs between the control and RCH adults of C. megacephala (fig. 4a), females had a greater transcriptional response than males and a greater transcriptional response was noted between the control and flies that had recovered from cold acclimation vs. differences noted between controls and flies that had only cold acclimated for 30 min (fig. 4). Specifically, 296 and 100 unigenes were differentially expressed between control and adults of R30F and R30M, respectively, including the up-regulation of 127 and 18 unigenes and down-regulation of 169 and 82 unigenes, respectively. Similarly, a total of 3053 and 1250 DEGs including 1761 and 753 up-regulated, 1292 and 753 down-regulated were detected comparing control and adults of RF and RM, respectively. Among these DEGs, females and males shared 32 unigenes between controls and R30F and R30M (fig. 4b), including two esterase genes (AchE), one collagen gene (COL3A), one Allantoinase gene (allB). Meanwhile, females and males shared 671 DEGs in response to 0°C for 2 h compared to controls (fig. 4e), and these unigenes generated eight different expression clusters (fig. 5a). The expression patterns of up-regulated genes shared by RF and RM are in Cluster 1, Cluster 3, Cluster 5, Cluster 6, and Cluster 8. These genes were enriched in Toll and Imd signaling pathway, spliceosome, protein processing in endoplasmic reticulum, MAPK signaling pathway, fat digestion and absorption, linoleic acid metabolism, and ether lipid metabolism (table 1). In the Toll and Imd pathways, nine genes were up-regulated, including three diptericin genes (Dpt) and two insect defensin genes (Def), ubiquitin-conjugating enzyme gene (Ube2N), fas-associated death domain protein (FADD), and two uncharacterized proteins. In the spliceosomal pathway HSP68, HSP70, HSP70A1, HSP70B, and two U6 snRNA-associated Sm-like protein genes (LSm2 and LSm8) were up-regulated. These HSP genes were also enriched in the MAPK signaling pathway. In addition, one growth arrest and DNA damage-inducible protein gene (GADD45) was up-regulated after acclimation that is involved in MAPK signaling pathway. Furthermore, six HSP70s, one HSP70-binding protein (HSP70BP), and four protein disulfide-isomerase genes (PDI) were enriched in protein processing in the endoplasmic reticulum gene set. Two up-regulated phospholipase genes (PLA2) were enriched in linoleic acid metabolism.

Figure 4. Analysis of differentially expressed genes (DEGs) for RCH in Chrysomya megacephala adults. (a) Distribution of up-regulated and down-regulated DEGs during the early stage and recovery stage of RCH and control in each comparison; (b–e) Venn diagram analysis of DEGs in different comparisons among groups. R30F, females during the early stage of RCH; R30M, males during the early stage of RCH; RF, females during the recovery stage of RCH; RM, males during the recovery stage of RCH; RCF, control females; RCM, control males.

Figure 5. Heatmap of differentially expressed genes (DEGs) involved in RCH and LCA in Chrysomya megacephala using an average value of three replicates. (a) Transcript abundance profiles and expression clusters of DEGs shared by female and male treated by 0°C for 2 h, prior to 25°C for 30 min. R30F, females during the early stage of RCH; R30M, males during the early stage of RCH; RF, females during the recovery stage of RCH; RM, males during recovery stage of RCH; RCF, control females; RCM, control males. (b) Transcript abundance profiles and expression clusters of DEGs shared by FTR and SCR. The treatments contain flies reared in fluctuating thermal regime (FTR) and stable cold regime (SCR) and control (LC) for 7 days. The order of cluster expression graph was based on the gene numbers of each cluster.
Table 1. KEGG pathways enriched in up-regulated genes shared by Chrysomya megacephala females and males during recovery stage of RCH (RF and RM)

DEGs of C. megacephala adults after LCA
To investigate gene expression that displayed significant changes during LCA, LCA-treated libraries were compared to the control to identify DEGs. We found a higher number of unigenes with differential expression for SCR than FTR. A total of 1993 up-regulated and 1526 down-regulated DEGs were detected for SCR, while 459 (up) and 231 (down) were detected for FTR (fig. 6a). In addition, FTR and SCR shared 453 DEGs (fig. 6b). There were six different expression clusters (fig. 5b). Cluster 1 and Cluster 3 showed the expression patterns of up-regulated genes in both FTR and SCR. These genes were significantly enriched in oxidative phosphorylation, glycerophospholipid metabolism, citrate cycle (TCA cycle), tight junction, amino sugar and nucleotide sugar metabolism, phototransduction – fly, regulation of actin cytoskeleton, Hippo signaling pathway – fly, Toll and Imd signaling pathway (table 2). There were 13 up-regulated genes involved in oxidative phosphorylation, including three ATP synthase genes (ATP5C1, ATP5O, ATP5D), one ATP synthase lipid-binding protein gene (ATP5G), one ATP synthase-coupling factor gene (ATP5J), five NADH dehydrogenase genes (NDUFB5, NDUFB6, NDUFB9, NDUFA11, ND5), one acyl carrier protein gene (NDUFAB1), one cytochrome b-c1 complex gene (QCR6), and one cytochrome b gene (CytB). Six esterase genes (AchE) were up-regulated and enriched in glycerophospholipid metabolism. Two isocitrate dehydrogenase genes (IDH), two pyruvate dehydrogenase genes (PDH), one malate dehydrogenase (MDH), and one citrate synthase gene (CS) were enriched in TCA cycle. Two chitinase genes and two chitin synthase genes were enriched in amino sugar and nucleotide sugar metabolism. One Rho-GTPase gene (RGTP), three actin genes, and one actinin gene were enriched in tight junction. Meanwhile these actin genes were involved in phototransduction – fly, regulation of actin cytoskeleton, and Hippo signaling pathway – fly. One Gram-negative bacteria-binding protein gene and two diptericin genes (Dpt) were up-regulated and enriched in Toll and Imd signaling pathway.

Figure 6. Analysis of differentially expressed genes (DEGs) for LCA in Chrysomya megacephala males. (a) Distribution of up-regulated and down-regulated DEGs of males exposed to fluctuating thermal regime (FTR) and stable cold regime (SCR) and control (LC) in each comparison; (b) Venn diagram analysis of DEGs in different comparisons.
Table 2. KEGG pathways enriched in up-regulated genes in Chrysomya megacephala shared by SCR and FTR

Validation of RNA-seq by qPCR
To validate the expression profiles identified from RNA-seq, we analyzed the transcript abundance of seven haphazardly selected DEGs using qPCR, including four up-regulated genes (fig. 7a–d) and three down-regulated genes (fig. 7e–g) for the recovery stage of RCH. qPCR showed that HSP40, HSP67, DIA1, and GADD45 were up-regulated significantly during recovery in both females and males (RF and RM). There was no significant difference between RCF and R30F, whereas the relative expressions of HSP40, HSP67, DIA1, and GADD45 in R30M were higher significantly than that of RCM (P < 0.05). The data from RNA-seq and qPCR were well correlated and fitted with a linear model y = 1.356x + 0.031 (R 2 = 0.819, P < 0.001) (fig. 7h).

Figure 7. Relative expression of cold stress-associated genes and validation of RNA-seq in RCH samples by qPCR. (a–g) Confirmatory expression profiles for seven genes identified in our transcriptomic screen for RCH by qPCR. *P < 0.05, ** means 0.001 < P < 0.01 (t-test). (h) Validation of RNA-seq by qPCR. R30F, females during the early stage of RCH; R30M, males during the early stage of RCH; RF, females during the recovery stage of RCH; RM, males during the recovery stage of RCH; RCF, control females; RCM, control males.
The selected seven genes had different expression patterns between FTR and SCR (fig. 8). There were two genes (DIA1 and GADD45) up-regulated in FTR but down-regulated in SCR (fig. 8a, b), and one gene (DPLX1) up-regulated in SCR but down-regulated in FTR (fig. 8e). Meanwhile, four genes (HSP40, HSP67, F2X2, SACX1) presented similar expression between FTR and SCR (fig. 8c, d, f and g). The data from RNA-seq and qPCR for LCA were well correlated and fitted with a linear model y = 1.155x + 0.078 (R 2 = 0.904, P < 0.001) (fig. 8h).

Figure 8. Relative expression of cold stress-associated genes and validation of RNA-seq in LCA samples by qPCR. (a–g) Confirmatory expression profiles for seven genes identified in our transcriptomic screen for RCH by qPCR. *P < 0.05, ** means 0.001 < P < 0.01, *** means <0.001 (t-test). (h) Validation of RNA-seq by qPCR. The samples contain males after FTR (fluctuating thermal regime :15/25°C for 7 days) and SCR (stable cold regime: reared in 15°C for 7 days), the adults reared in 25°C for 7 days as control.
Discussion
Low temperatures in early spring inhibit insect growth, survival, and activity (Gruner et al., Reference Gruner, Slone, Capinera and Turco2017), thus there are few pollinators for early spring crops like oilseed rape and strawberry. Chrysomya megacephala and its closely related species are potentially important pollinator species (Wang et al., Reference Wang, Zhao and Lei2005; Yanagi et al., Reference Yanagi, Miura, Isobe, Okuda and Yoshida2017). Thus our goal was to better understand the process of cold acclimation in this species as a starting point for developing protocols that could be used to improve pollinator performance on early season crops. We imagine that programs for mass rearing and acclimating large numbers of C. megacephala then releasing them to pollinate in the early season when other pollinators are rare could improve agricultural yields (Wang et al., Reference Wang, Zhao and Lei2005). In this study, C. megacephala adults pre-exposed to 0°C for 1–4 h had increased survival of a 2 h cold shock at −6°C compared to unacclimated controls. Such phenotypic plasticity has been reported in many insects. For example, the survival rate of S. crassipalpis and Lissorhoptrus oryzophilus pre-treated by RCH was significantly increased when exposed to lethal cold temperature (Lee et al., Reference Lee, Chen and Denlinger1987; Yang et al., Reference Yang, Zhang, Wang, Wang, Pan, Zhang and Xi2018). Likewise, LCA (at either constant or fluctuating regimes) increased survival after cold shock at −6°C for 2 h and decreased the recovery time from cold exposure to 0°C that induced chill coma. These results indicate that cold-acclimated C. megacephala could survive at lower temperatures (during cold storage or releasing in the field) and recover faster when the temperature rises gradually (night to day in the field). Our results are also consistent with the work in other systems. For example, cold acclimation enhances flight performance of T. leucotreta under low-temperature conditions in the field (Boersma et al., Reference Boersma, Boardman, Gilbert and Terblanche2019), and cold-acclimated D. melanogaster have a stronger ability to find resources in the cold in the field (Kristensen et al., Reference Kristensen, Hoffmann, Overgaard, Sorensen, Hallas and Loeschcke2007). Therefore, future research should focus on whether cold-acclimated C. megacephala adults have stronger flight ability and the ability to locate food than non-cold-acclimated flies in the early spring, which would improve pollination efficiency of early spring crops.
Transcriptomics, proteomics, and metabolomics have been used to explore the mechanisms of RCH and LCA (Teets et al., Reference Teets, Peyton, Ragland, Colinet, Renault, Hahn and Denlinger2012; MacMillan et al., Reference MacMillan, Knee, Dennis, Udaka, Marshall, Merritt and Sinclair2016; Shearer et al., Reference Shearer, West, Walton, Brown, Svetec and Chiu2016; Teets and Denlinger, Reference Teets and Denlinger2016; Enriquez et al., Reference Enriquez, Renault, Charrier and Colinet2018). In this study, we performed transcriptomics/RNAseq to study different molecular mechanisms of RCH and LCA in C. megacephala for the first time. The sequencing and assembly results provided a deeply improved and comprehensive transcriptome database of C. megacephala adults over a previous study (Zhang et al., Reference Zhang, Yu, Yang, Song, Hu and Zhang2013; Wang et al., Reference Wang, Xiong, Lei and Zhu2015b). Therefore, our data provide valuable information for future investigation of the molecular mechanisms of cold tolerance in C. megacephala or other species and facilitate future evolutionary studies and comparative genomic studies in C. megacephala.
During the early stage of RCH, after only 30 min of exposure, females and males shared only 32 DEGs. There was no pathway enriched significantly by analyzing up-regulated genes during this early stage of RCH. Thus, RCH may be primarily mediated by second messenger systems, such as protein phosphorylation or calcium accumulation, with fewer gene products required (Fujiwara and Denlinger, Reference Fujiwara and Denlinger2007; Teets et al., Reference Teets, Yi, Lee and Denlinger2013). After RCH, Toll and Imd signaling pathways were enriched, which is consistent with other insects under cold stress, e.g. D. melanogaster and Sogatella furcifera (Huang et al., Reference Huang, Xue, Zhuo, Cheng, Xu and Zhang2017; Stetina et al., Reference Stetina, Poupardin, Moos, Simek, Smilauer and Kostal2019). Temperature stress appears to have activated immunity before the pathogen elicited a response from the insect, and this early activity prevented infection under stressful conditions (Xu and James, Reference Xu and James2012). However, it will require further effort to learn whether transcriptional activation of Toll and Imd signaling pathway and diptericin synthesis can benefit stress and immune responses in this insect. HSPs are well-known chaperone proteins that play important roles in thermal resistance and organismal responses to stressors (starvation, heavy metals) (Wang et al., Reference Wang, Li, Zhu, Fang and Ye2012). Several HSPs including HSP40, HSP67B, and HSP70 were up-regulated in C. megacephala females and males during the recovery stage of RCH. Meanwhile, these HSPgenes and up-regulated Sm-like protein genes were enriched in the spliceosome pathway that is important to regulating transcription and the products eventually emerging from translation. Previous studies indicated that spliceosomal activity is induced by multiple stress conditions (Dutertre et al., Reference Dutertre, Sanchez, Barbier, Corcos and Auboeuf2011; Li et al., Reference Li, Fu, Qu, Wang, Xu and Luo2017). For example, in D. melanogaster, alternative splicing regulates the transcriptional activity of heat shock transcription factor in response to cold/heat stress (Fujikake et al., Reference Fujikake, Nagai, Popiel, Kano, Yamaguchi and Toda2005). Therefore, cold-induced genes related to the spliceosome may play important roles in increasing the survival of C. megacephala at low temperatures. Low temperatures threaten the stability and function of proteins, and cold tolerance can result from enhanced protein protection or repair (refolding) of denatured or misfolded proteins (Bhatnagar et al., Reference Bhatnagar, Bogner and Pikal2007). In our study, many up-regulated genes (containing HSP70, HSP70BP, and DPI) involved in protein processing in the endoplasmic reticulum were enriched in C. megacephala after RCH. These proteins could help C. megacephala protect and repair critical proteins during and after exposure to damaging low temperatures. Overall, C. megacephala adults appear to activate their immune response, spliceosome, and protection and repair of proteins to increase cold tolerance for RCH.
To evaluate the effects of longer term cold acclimation on C. megacephala, fluctuating acclimation (FTR) and stable cold acclimation (SCR) treated flies were compared to flies kept at stable warm temperatures. Although, there were some differences between the transcripts associated with the two acclimation regimes, most DEGs of FTR and SCR had similar expression patterns, similar results have been observed in D. melanogaster (Sørensen et al., Reference Sørensen, Schou, Kristensen and Loeschcke2016). Therefore, KEGG pathway enrich was carried out using genes up-regulated in both FTR and SCR. Up-regulated genes involved with oxidative phosphorylation were highly enriched in cold-acclimated C. megacephala, while the oxidative phosphorylation pathways were inhibited in some species treated by cold (Nylin, Reference Nylin2013; Cui et al., Reference Cui, Hu, Wang, Tao and Zong2017). Genes related to oxidative phosphorylation were up-regulated, presumably to compensate for lowered enzyme efficiency at lower temperatures. Up-regulated ATP synthase genes could give higher metabolic rates in the cold, which could produce heat to raise body temperatures and provide energy for greater activity in the cold. Six genes related to the TCA cycle were up-regulated, which also may indicate that C. megacephala can maintain energy production in the cold. With respect to amino sugar and nucleotide sugar metabolism, chitinase genes and chitin synthase genes were up-regulated. The fact that both catabolic and anabolic enzymes are up-regulated may suggest a high rate of chitin turnover. The phenomenon that chitin synthase genes up-regulated has also been observed in the winter morphs of D. suzukii (Shearer et al., Reference Shearer, West, Walton, Brown, Svetec and Chiu2016). These results indicated that chitin could play an important role in cold tolerance of insects, but more functional work on chitin metabolism is needed to test this hypothesis. In our study, cold-acclimated C. megacephala had altered expression of cytoskeletal branching and stabilizing genes such as actin, RGTP, and actinin, suggesting that cold acclimation enhanced the stabilization of cytoskeleton in cold, consistent with previous report in G. pennsylvanicus (Kim et al., Reference Kim, Robich, Rinehart and Denlinger2006; Des Marteaux et al., Reference Des Marteaux, McKinnon, Udaka, Toxopeus and Sinclair2017). Overall, transcriptomic analysis showed that carbohydrate metabolism pathways containing TCA cycle, and oxidative phosphorylation were altered in cold acclimation. Meanwhile, we hypothesize that C. megacephala improved the stability of cells and tissues by altering actin and chitin after LCA.
To compare the RCH and LCA in a holistic perspective, we analyzed metabolic pathways using iPath. Compared with RCH (fig. S4), LCA showed a broader signal of metabolic regulation in mRNA levels based on the DEG distribution (fig. S5). Earlier studies have suggested that RCH is primarily a cell-mediated event with little or no reliance on transcription, while LCA appears to involve dramatic changes in the transcriptome (Teets et al., Reference Teets, Peyton, Ragland, Colinet, Renault, Hahn and Denlinger2012; Teets and Denlinger, Reference Teets and Denlinger2013). This has some implications for the perspective of using these treatments, for example, both treatments improve cold tolerance of C. megacephala adults thus RCH might be very effective as it only triggers fewer genes and affects the general metabolisms less. However, the RCH response might be less permanent (i.e. disappear faster) so there is much more to investigate regarding the survival, locomotion, and pollination efficiency of the insects after field released.
Conclusion
We showed that both RCH and LCA can improve cold hardiness in C. megacephala. In our study, transcriptomics revealed candidate mechanisms for RCH and LCA responses in C. megacephala. Many genes related to immune response, the spliceosome, and protein processing in the endoplasmic reticulum were up-regulated during the recovery stage of RCH. In contrast, LCA was associated with carbohydrate metabolism (TCA cycle and oxidative phosphorylation), and cytoskeleton branching and stabilizing. There were more genes and pathways enriched in LCA than RCH in C. megacephala. The partly independent molecular responses to RCH and LCA suggest that several avenues for manipulating cold performance exist in this species, and RCH may be better method for pollination application of cold-acclimated C. megacephala due to shorter acclimation duration and less metabolic adjustment than LCA. These results lay a foundation for identifying mechanisms underlying RCH and LCA, as well as offering a promising approach for increasing cold tolerance of beneficial insects and extending their applications in the near future.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485321001073
Acknowledgements
This study was funded by the National Natural Science Foundation of China (31972270, 31661143045), Agricultural public welfare industry research from the Ministry of Agriculture of People's Republic of China (201503137), and the Coordinated Research Project (18269) from the International Atomic Energy Agency. We are grateful to Jesper Givskov Sørensen and Hahn Daniel Allen for their kindly help during manuscript preparation.
Conflict of interest
None.













