Introduction
It is generally considered that alien insect parasitoids can have positive effects on the environment and the economy (Roy et al., Reference Roy, Roy and Roques2011). There are more than 200 recorded alien hymenopteran parasitoid species in Europe. Unlike herbivorous or predatory insects, most parasitoids were intentionally introduced as biocontrol agents (BCAs) (Rasplus et al., Reference Rasplus, Villemant, Paiva, Delvare and Roques2010). Braconidae is one of the largest hymenopteran families in Europe, with 3500 recorded species (van Achterberg, Reference Van Achterberg2013), among which 18 are considered to be alien species (Rasplus et al., Reference Rasplus, Villemant, Paiva, Delvare and Roques2010; Petrović et al., Reference Petrović, Mitrović, Starý, Petrović-Obradović, Tomanović, Žikić and Vorburger2013, Reference Petrović, Čkrkić, Jamhour, Petrović-Obradović, Mitrović, Starý, Nedstam and Tomanović2017). However, the poorly known taxonomy of parasitoids has frequently resulted in overlooked alien parasitoid species (Ye et al., Reference Yu, van Achterberg and Horstmann2017). Seven out of 18 alien braconid species belong to the subfamily Aphidiinae. Species Aphidius colemani Viereck, Aphidius smithi Sharma & Subba Rao, Lysiphlebus testaceipes Cresson, Pauesia cedrobii Starý & Leclant and Pauesia unilachni Gahan were introduced intentionally as BCAs (Rasplus et al., Reference Rasplus, Villemant, Paiva, Delvare and Roques2010; Roy et al., Reference Roy, Roy and Roques2011). On the other hand, species Lysiphlebus orientalis Starý & Rakhshani and Aphidius ericaphidis Pike & Starý were accidentally introduced to Europe (Petrović et al., Reference Petrović, Mitrović, Starý, Petrović-Obradović, Tomanović, Žikić and Vorburger2013, Reference Petrović, Čkrkić, Jamhour, Petrović-Obradović, Mitrović, Starý, Nedstam and Tomanović2017). Most of the aphidiine BCAs were introduced to Europe in the period from the 1930s to the 1980s, when biocontrol programmes were less controlled in terms of monitoring the non-target effects of released agents (Rasplus et al., Reference Rasplus, Villemant, Paiva, Delvare and Roques2010). Even today, it is not common practice to look for some ecological aspects of potential BCAs such as displacement of native species through competition (Bennett, Reference Bennett1993). However, there are examples of aphidiine parasitoids being displaced by L. testaceipes in Europe (see Žikić et al., Reference Žikić, Stanković, Ilić-Milošević, Petrović-Obradović, Petrović, Starý and Tomanović2015) or by Aphidius ervi Haliday in North America (Schellhorn et al., Reference Schellhorn, Kuhman, Olson and Ives2002). Consequently, the ecological costs of biological control are most likely underestimated. Apart from the gaps in knowledge about ecology, there is an evident problem with the taxonomy of some aphidiine BCAs. A recent taxonomic study on the A. colemani species group has shown that it consists of three species instead of two and that commercially cultured and distributed BCAs tend to be a mixture of all three species (Tomanović et al., Reference Tomanović, Petrović, Mitrović, Kavallieratos, Starý, Rakhshani, Rakhshanipour, Popović, Shukshuk and Ivanović2014), although there are evident differences in their ecological properties (e.g., host preference (Kavallieratos & Lykouressis, Reference Kavallieratos and Lykouressis1999; Lozier et al., Reference Lozier, Roderick and Mills2009)).
The pea aphid (Acyrthosiphon pisum Harris) and blue alfalfa aphid (Acyrthosiphon kondoi Shinji) are species of high agricultural importance with worldwide distribution (van Emden & Harrington, Reference van Emden and Harrington2007). Acyrthosiphon pisum is a pest of many herbaceous and shrubby legumes, including some of economic importance like peas and alfalfa worldwide (Blackman & Eastop, Reference Blackman and Eastop2000). Acyrthosiphon kondoi has an Asian origin, but it is widely spread in the Americas, Africa and the Australian region (Berberet et al., Reference Berberet, Arnold and Soteres1983). Recently, it was also recorded in Europe (Coeur d'acier et al., Reference Coeur d’acier, Pérez Hidalgo, Petrović-Obradović, Roques, Kenis, Lees, Lopez-Vaamonde, Rabitsch, Rasplus and Roy2010). Aphidiine parasitoids have been used in biological control programmes against A. pisum and A. kondoi since the late 1950s all over the world (Mackauer & Finlayson, Reference Mackauer and Finlayson1967; Starý, Reference Starý1974; González et al., Reference González, Hagen, Starý, Bishop, Davis, Pike, Nechols, Andres, Beardsley, Goeden and Jackson1995; Summers, Reference Summers1998; Wylie et al., Reference Wylie, Matheson, Uddin and Holliday2005). Apart from A. ervi, species from the Aphidius eadyi group are most commonly used in those biocontrol programmes. The A. eadyi species group can be defined as a group of species with a costulate anterolateral area of the petiole that parasitize A. pisum. It consists of three species: A. smithi, A. eadyi Starý, González & Hall and Aphidius banksae Kittel ( = A phidius staryi sens. auct – Kittel, Reference Kittel2016).
Aphidius smithi was first identified in 1958 as being one of the main species responsible for natural control of the pea aphid in India. Even prior to its description, the new species had been released in California alfalfa fields, where in 1 year it became established and provided considerable control of the pea aphid (Hagen & Shlinger, Reference Hagen and Shlinger1960). After this initial success, mass releases were continued in North America during the 1960s and 1970s. Meanwhile, there were a few experimental releases of A. smithi in Europe (in Poland, the Czech Republic and Moldavia) (see Starý, Reference Starý1974), but initial results showed that there were no establishments of parasitoid populations (Starý, Reference Starý1974). The appearance of the new aphid pest A. kondoi in the Americas, Australia and New Zealand prompted new research efforts to find additional BCAs throughout the Old World (González et al., Reference González, White, Hall and Dickson1978; Starý et al., Reference Starý, González and Hall1980). Those investigations resulted in the description of A. eadyi, which is widely distributed throughout the Palaearctic (Starý et al., Reference Starý, González and Hall1980). The description of A. eadyi, member of the Aphidius urticae group, sheds some light on this taxonomically problematic group of species (Starý et al., Reference Starý, González and Hall1980). During the 1970s, there was in fact a deep gap in the knowledge about taxonomy of the A. urticae species group, whose members also attack the pea aphid and were probably introduced to the USA for biocontrol purposes (Marsh, Reference Marsh1977). Starý et al. (Reference Starý, González and Hall1980) concluded that most of the parasitoid specimens attacking pea aphids identified as A. urticae Haliday or members of the A. urticae species group were actually A. eadyi. As a specific parasitoid of the pea aphid, A. eadyi was introduced as a BCA in Burundi and New Zealand (Cameron et al., Reference Cameron, Walker and Allan1981; Autrique et al., Reference Autrique, Starý and Ntahimpera1989; Cameron & Walker, Reference Cameron and Walker1989), where it established stable populations 1 year after release and succeeded in reducing pea aphid populations (Cameron et al., Reference Cameron, Walker and Allan1981). Research projects on biocontrol of the pea aphid in North America resulted in the description of one more species from the A. eadyi group – A. staryi Chen & Luhman (Chen et al., Reference Chen, González and Luhman1990). In 1983 and 1984, pea aphid parasitoids were collected from Israel and Turkey and introduced to the USA as A. smithi. Later on it was shown that those specimens differed from true A. smithi specimens in morphology, biology and isozyme patterns (Unruh et al., Reference Unruh, White, González and Woolley1989; Chen et al., Reference Chen, González and Luhman1990) and they were described as A. staryi (Chen et al., Reference Chen, González and Luhman1990). However, it turned out that A. staryi is a primary junior homonym, and its name was changed to A. banksae (Kittel, Reference Kittel2016). After the description of A. banksae and its introduction to California (González et al., Reference González, Hagen, Starý, Bishop, Davis, Pike, Nechols, Andres, Beardsley, Goeden and Jackson1995), the species was forgotten. In the literature, A. banskae was mentioned only a few times after its description (Atanassova, Reference Atanassova1997; Atanassova et al., Reference Atanassova, Brookes, Loxdale and Powell1998; Akar & Çetin Erdoğan, Reference Akar and Çetin Erdoğan2017). Although exhausting efforts have been made to control A. pisum with aphidiine wasps, there is still a gap in knowledge and confusion about its parasitoid spectrum, especially in Europe. Aphidius urticae still represents the biggest source of confusion. There have been a few attempts to clarify the status of members of the A. urticae species group feeding on pea aphids (Starý, Reference Starý1974; Starý et al., Reference Starý, González and Hall1980; Chen et al., Reference Chen, González and Luhman1990), but the group's overall taxonomic status remains unclear. Species of this group have been repeatedly reported throughout Europe (Atanassova, Reference Atanassova1997; Kavallieratos et al., Reference Kavallieratos, Tomanović, Starý, Athanassiou, Sarlis, Petrović, Niketić and Anagnou Veroniki2004; Alhmedi et al., Reference Alhmedi, Haubruge and Francis2009; Žikić et al., Reference Žikić, Ilić-Milošević, Stanković, Petrović, Petrović-Obradović, Kavallieratos, Starý and Tomanović2012; Derocles et al., Reference Derocles, Plantegenest, Rasplus, Marie, Evans, Lunt and Le Ralec2016). Additional doubts about the identity of members of the A. urticae species group from pea aphids were aroused by Derocles et al. (Reference Derocles, Plantegenest, Rasplus, Marie, Evans, Lunt and Le Ralec2016), who found significant genetic differences between specimens from Microlophium carnosum (Buckton) and A. pisum, and by Jamhour et al. (Reference Jamhour, Mitrović, Petrović, Starý and Tomanović2016), who stated that speciation in this group is driven by specialization to different aphid hosts.
The aim of the present study was to employ molecular tools and geometric morphometrics to resolve the taxonomic status of BCAs belonging to the A. eadyi species group. We here explore and discuss the genetic structure and morphological variability of members of the A. eadyi group from a wide area of distribution. In addition, A. banksae is redescribed, diagnosed and illustrated.
Material and methods
Insect material
Samples were collected worldwide over the last 40 years (table 1). Some of the insect material was obtained from field sampling of plant parts (leafs and stems) infested by aphids and reared until emergence of parasitoids. Other specimens were obtained by rearing in insectaries of parasitoids collected for biocontrol programmes by Professor Dan González during his field trips in Asia trying to obtain suitable aphid parasitoid species for their introduction to alfalfa fields in the USA.
Table 1. List of specimens belonging to Aphidius eadyi group, reared from A. pisum, submitted to molecular and morphometric analyses.

1 Origin of populations reared in insectaries at the University of California, Riverside, CA, USA.
2Specimens designated with same sign are reared from same aphid population – sample.
3 Sequences retrieved from GenBank.
DNA extraction, PCR amplification and sequencing
DNA was extracted from entire wasp specimens (42 specimens in total) using the KAPA Express Extract kit (Kapa Biosystems Inc., Boston, MA, USA) or the Dneasy® Blood & Tissue Kit (Qiagen Inc., Valencia, CA, USA) following the manufacturer's instructions. The barcoding region of the cytochrome oxidase subunit I mitochondrial gene (mtCOI) was amplified using two different methods, depending on the sample's condition. DNA extracted from collected specimens preserved in 96% ethanol was amplified using the standard barcoding primers LCO1490 and HCO2198 (Folmer et al., Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994). Amplification was performed in a final volume of 50 µl. The reaction mixture contained 2 µl of extracted DNA as the template, 1× KAPA2G Robust HotStart ReadyMix (containing 2 mM MgCl2 at 1×) (Kapa Biosystems Inc.), and 0.5 µM of each primer. All PCR reactions were conducted in an Eppendorf Mastercycler® (Hamburg, Germany) using a thermal profile taken from Petrović et al. (Reference Petrović, Mitrović, Starý, Petrović-Obradović, Tomanović, Žikić and Vorburger2013). Mitochondrial COI barcoding fragments from dry material could not be amplified with standard LCO1490/HCO2198 primers. In such cases, the Aphidius-specific degenerative primers Aph1Rd, Aph2Fd, AphsRd and Aph3Fd were used to amplify short fragments following specific direct and nested PCR protocols (Jamhour, Reference Jamhour2017; Mitrović & Tomanović, Reference Mitrović and Tomanović2018), and thereafter aligned to a complete sequence for further comparison. DNA sequencing was performed by Macrogen Inc. (Seoul, Korea). The nucleotide sequence data were deposited in the GenBank database under accession numbers MG987145–MG987170.
Genetic analysis
Sequence editing was performed using FinchTV (http://www.geospiza.com), while CLUSTALW (Thompson et al., Reference Thompson, Higgins and Gibson1994) integrated in MEGA6 software was used for sequence alignment (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013). Average genetic distances between sequences within each group and between the groups of species were calculated using Kimura's two-parameter method (K2P) of base substitution (Kimura, Reference Kimura1980). Phylogenetic trees were obtained using the maximum likelihood (ML), maximum parsimony (MP) and neighbour joining (NJ) methods integrated in MEGA6 software. For all methods, 1000 bootstrap replicates were performed to assess the branch support. In the case of ML, the best-fitting model of sequence evolution based on the Bayesian Information Criterion and Akaike Information Criterion corrected (Nei & Kumar, Reference Nei and Kumar2000) was the Hasegawa–Kishino–Yano model (Hasegawa et al., Reference Hasegawa, Kishino and Yano1985) with invariant sites (HKY + I), as determined by Modeltest (Posada & Crandall, Reference Posada and Crandall1998). The sequence of Areopraon chaitophori (GenBank Acc. No. KC128679) was used as an outgroup for phylogenetic analyses. An A. banksae haplotype network based on statistical parsimony with a confidence limit of 95% was created using the TCS programme, ver. 1.21 (Clement et al., Reference Clement, Posada and Crandall2000).
In addition, two methods of DNA taxonomy were used to identify species from COI sequence data. First, we used the Poisson Tree Process (PTP) that identified genetic clusters representing independently evolving entities, optimizing differences in branching patterns within and between taxa (Zhang et al., Reference Zhang, Kapli, Pavlidis and Stamatakis2013). PTP was applied on MP tree using its online tool (http://species.h-its.org/ptp/) with default settings. In our data set, there is uneven sampling of individuals per species and there was a possibility that PTP method could overestimate the number of recognized species (Zhang et al., Reference Zhang, Kapli, Pavlidis and Stamatakis2013). Automatic Barcode Gap Discovery (ABGD) method was used in order to validate previous methods. ABGD tests the existence of a barcode gap in genetic distances and then identifies species as groups of individuals united by shorter genetic distances than the gap (Puillandre et al., Reference Puillandre, Lambert, Brouillet and Achaz2012). ABGD was applied on COI alignment through its online tool (http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html) using the K2P of pairwise distances (Kimura, Reference Kimura1980).
Geometric morphometrics
Variation in wing size and shape was analysed using the geometric morphometric approach. Female right forewings were used for this purpose. In total, 223 individuals from 13 geographically distant populations belonging to the A. eadyi species group (A. banksae – 46 specimens; A. eadyi – 87 specimens; A. smithi – 90 specimens; table 1) were dissected, slide mounted and analysed. All wings were photographed under the same magnification and microscope settings. A constellation of 13 homologous landmarks (fig. 1) was used in order to explore and quantify the variation of wing size and shape.

Fig. 1. Landmarks scored on the right forewing of an Aphidius eadyi female.
Generalized Procrustes Analysis (Rohlf & Slice, Reference Rohlf and Slice1990; Dryden & Mardia, Reference Dryden and Mardia1998) was applied to obtain a matrix of wing shape coordinates (Procrustes coordinates) from which all differences due to position, scale and orientation had been discarded. Wing size was computed as the centroid size (CS), which is the measure of size in geometric morphometrics and reflects the amount of dispersion around the centroid of landmark configuration (see Zelditch et al., Reference Zelditch, Swiderski and Sheets2012). To explore the variation in wing shape, we carried out a multivariate ordination by Principal Component Analysis (PCA) based on the covariance matrix. The differences in the wing size (CS) and the wing shape between phylogenetic lineages were tested with analysis of variance (ANOVA) and multivariate analysis of variance (MANOVA), respectively. In order to test whether species of Aphidius wasps can be distinguished on the basis of forewing morphology, we conducted a discriminant analysis of pairwise Procrustes distances between forewings of the phylogenetic lineages/species studied. The reliability of species identification was assessed by Discrimination function analysis and through cross-validation (Lachenbruch, Reference Lachenbruch1967). Wing shape changes were visualized by creating outline-wrapped graphs.
To reconstruct and visualize evolutionary shape changes, we mapped the obtained PC scores onto the phylogeny. Shapes corresponding to the internal nodes were reconstructed using the weighted squared-change parsimony method (Maddison, Reference Maddison1991; Klingenberg & Gidaszewski, Reference Klingenberg and Gidaszewski2010). Statistical significance of differences in wing shape between species was obtained by permutation test (10,000 permutations). The ellipses are drawn and sized to comprise 90% of the observations referring to the three phylogenetic lineages/species.
All analyses were performed using MorphoJ software (Klingenberg, Reference Klingenberg2011), except for ANOVA, MANOVA and the Tukey HSD test, which were conducted with SAS, ver. 9.1.3 (SAS Institute Inc., Cary, NC, USA). The herein employed morphological terminology relating to diagnostic characters of the parasitoid species is based on Sharkey & Wharton (Reference Sharkey, Wharton, Wharton, Marsh and Sharkey1997).
Results
Genetic relationships
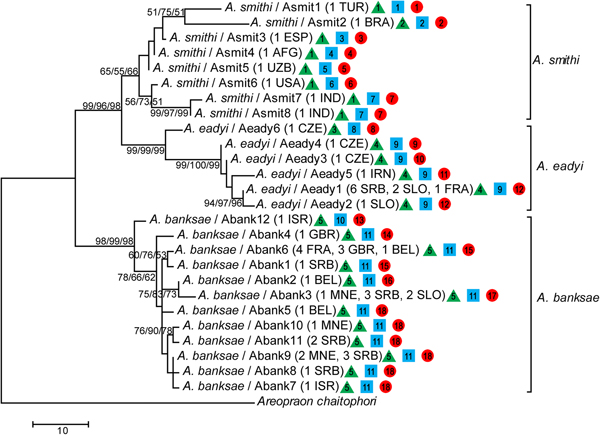
In total, 51 barcoding fragments of the mtCOI gene were amplified and submitted to analyses and reconstruction of phylogenetic relationships between species belonging to the A. eadyi group. All obtained phylogenetic trees (ML, MP and NJ) showed the same topology, where specimens belonging to species A. banksae, A. eadyi and A. smithi form separate clusters (fig. 2).
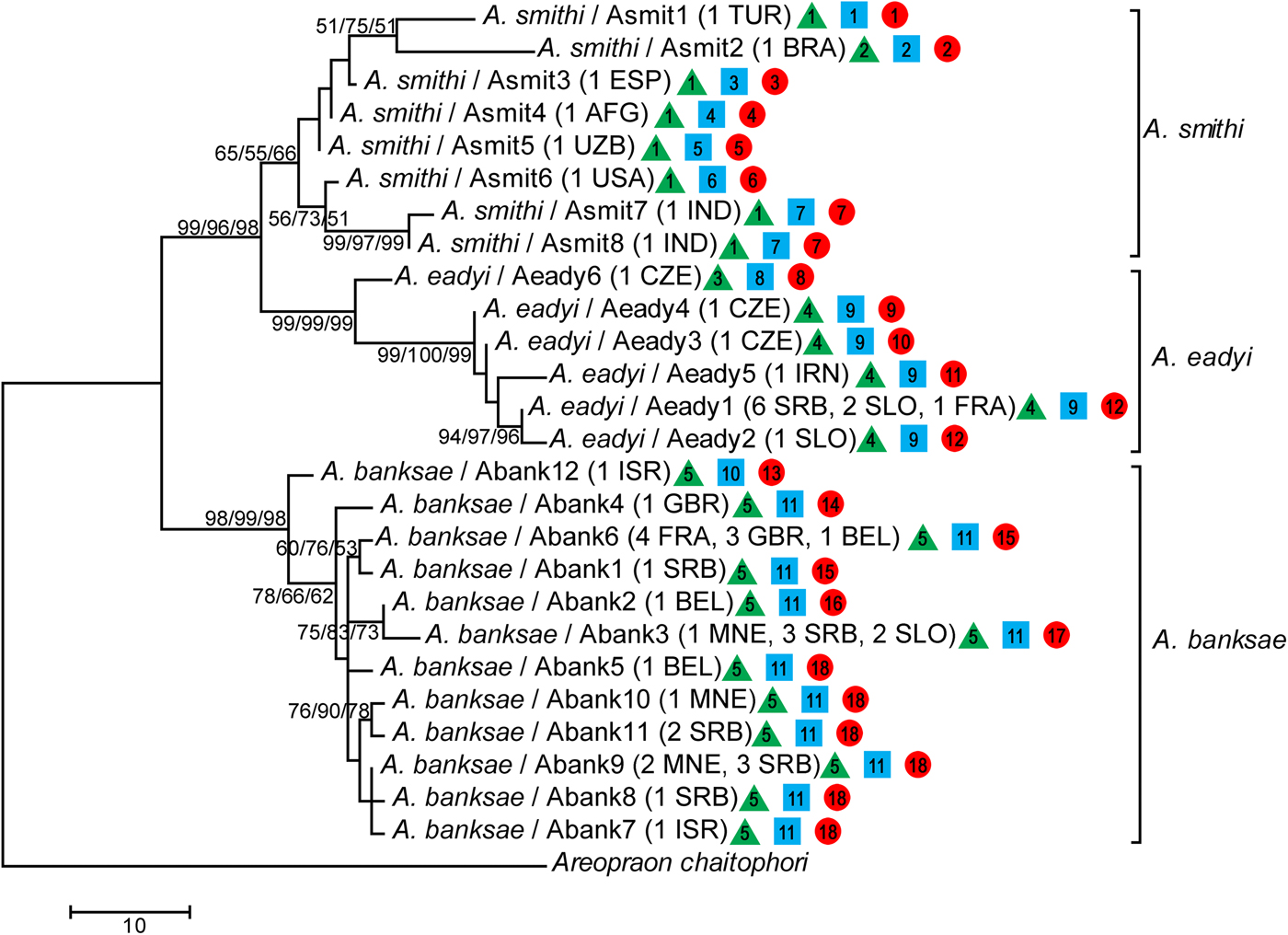
Fig. 2. Phylogenetic tree based on partial mtCOI sequences obtained using maximum likelihood (ML), maximum parsimony (MP) and neighbour-joining (NJ) methods. Bootstrap values are indicated above/below branches in the following order: ML/MP/NJ. Numbers and letters in parentheses refer to the number of sequences for each haplotype and geographic origin of sequences, respectively. The assignment of specimens to potential independently evolving entities is presented by green triangle (ABGD), blue square (PTP ML) and green circle (PTP BI) where different numbers represent different entities.
The species A. banksae and A. eadyi were clustered as separate taxa with very strong bootstrap support (>95%), while clustering of A. smithi can be treated as unsupported with bootstrap values of ~60%. Moreover, A. eadyi and A. smithi clustered together with >96% bootstrap support, which corresponds with a lower average K2P genetic distance between these two taxa in comparison with A. banksae (table 2). We determined the existence of 26 different haplotypes in total (A. banksae: Abank1–12, A. eadyi: Aeady1–6, A. smithi: Asmit1–8).
Table 2. Mean K2P genetic distances between (italic) and within the groups of parasitoids belonging to the Aphidius eadyi group.

Each of the eight individuals of A. smithi showed a different COI haplotype. The K2P genetic distances between A. smithi haplotypes was surprisingly high, ranging from 0.2% between Asmit4 (from Afghanistan) and Asmit5 (Uzbekistan) up to 4.3% between Asmit1 and Asmit7 from Turkey and India, respectively. A total of 14 A. eadyi specimens were included in molecular analysis, which resulted in identification of six haplotypes (Aeady1–6) with a mean K2P genetic distance of 1.5%. The most diverged haplotype was Aeady6, which was identified in one specimen from the Czech Republic. Genetic distances between Aeady6 and the other A. eadyi haplotypes ranged from 2.3 to 3.4%, and it even forms its own phylogenetic clade (fig. 2). Haplotypes Aeady1–5 differ from each other in the range of 0.2–1.4%. The most common haplotype in the analysed material was Aeady1, which was identified in nine specimens originating from France, Serbia and Slovenia. All other haplotypes (Aeady2–5) were identified in a single specimen. The highest number (12) of haplotypes was detected in 30 analysed A. banksae specimens (Abank1–12). The mean K2P genetic distance between A. banksae haplotypes was 1%. The most dominant haplotype was Abank6, which was determined in eight specimens, followed with Abank3 and Abank9, identified in six and five specimens, respectively (table 1, figs 2 and 3).
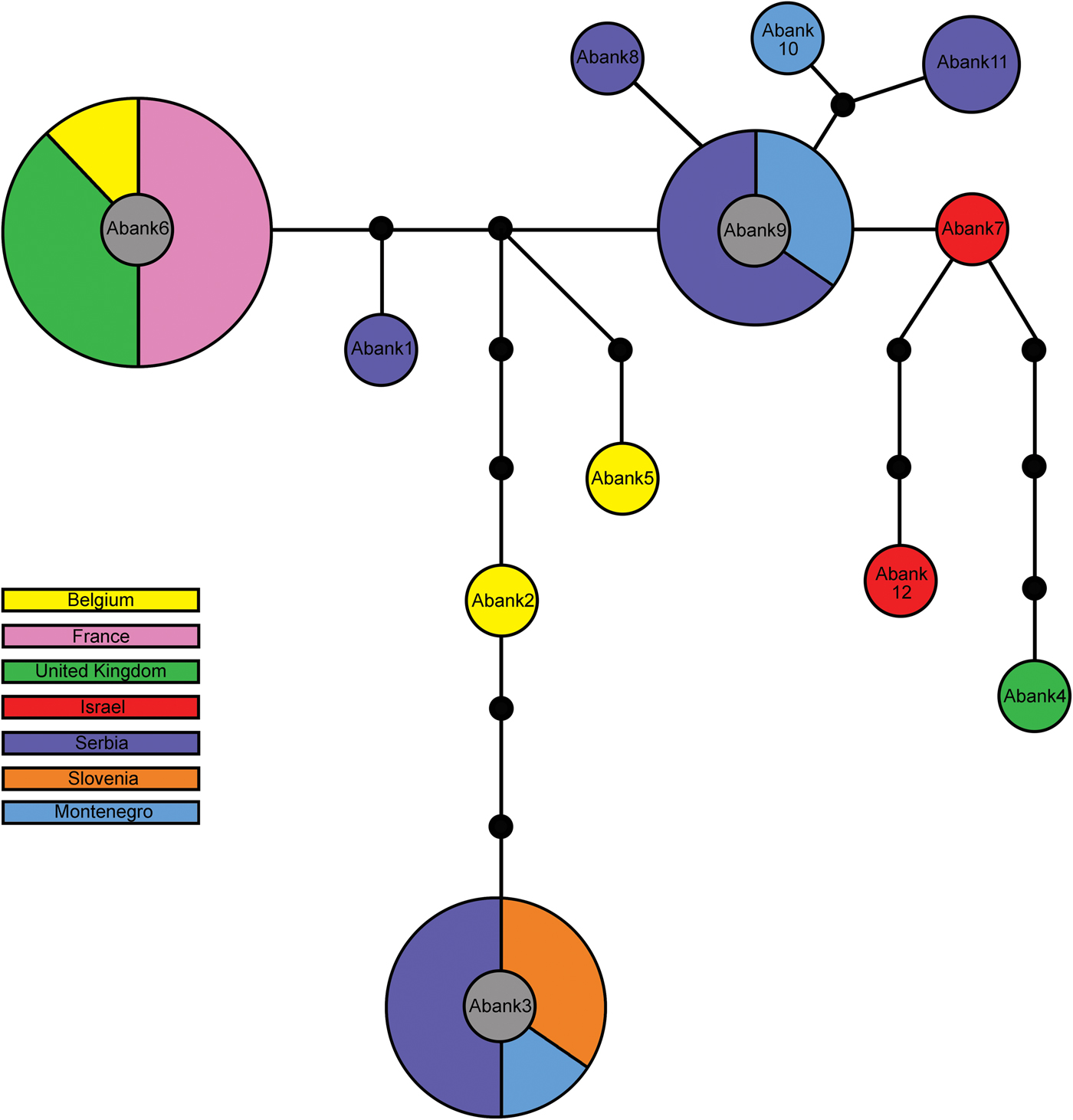
Fig. 3. Haplotype network obtained from 30 Aphidius banksae specimens using statistical parsimony (TCS). Circles represent specific haplotypes, size of the circle reflecting the number of individuals with that haplotype (not to scale), the colour its geographic distribution. Smaller filled circles represent missing haplotypes; lines between circles are mutational steps.
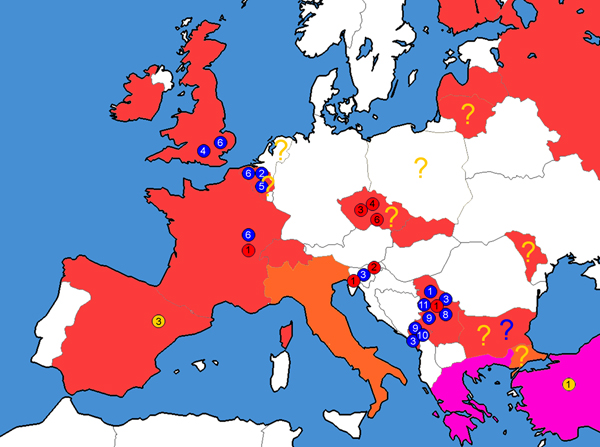
Aphidius banksae was previously overlooked in Europe, and we here determine that it is present and widely distributed from the UK in the west to the Balkans in the east (table 1, figs 2–4).

Fig. 4. Distribution of analysed haplotypes of the Aphidius eadyi group in Europe. Circles in different colours and numbers represent different haplotypes, while colour on the map represents distribution of the species adapted after van Achterberg (Reference Van Achterberg2013). Colour code: red – A. eadyi; yellow – A. smithi; blue – A. banksae; orange – A. eadyi and A. smithi co-occurring; pink – A. banksae, A. eadyi and A. smithi co-occurring.? – Literature finding to be taken with caution.
Both species discovery methods revealed genetic discontinuities that might indicate independently evolving lineages within A. smithi and A. eadyi. The PTP method identified 11 taxa based on ML solutions (PTP ML), and 18 taxa based on Bayesian solutions (PTP BI) (fig. 2). Both PTP ML and PTP BI identified highest number (seven) of hidden taxa within A. smithi. PTP ML identified additional two taxa within A. eadyi as well as within A. banksae, while PTP BI identified five taxa within A. eadyi and six taxa within A. banksae (fig. 2). Although the number of species was possibly overestimated, there are similarities with ABGD results. ABGD method provided estimate of five taxa in total, separating haplotypes Asmit2 (within A. smithi) and Aeady6 (within A. eadyi) as independently evolving lineages. Haplotypes Asmit2 and Aeady6 were recognized as separate entities by both species discovery methods as well as by genetic distances and phylogeny.
Geometric morphometrics
Species of the A. eadyi group do not differ in forewing size (one-way ANOVA, F 2, 220 = 0.903; P = 0.407). All three species differ significantly in forewing shape (MANOVA, Wilks’ λ = 0.25413, F 44, 398 = 8.90; P < 0.0001).
The first three axes (PC1, PC2 and PC3) describe 54.3% of total variance in wing shape (with PC1, PC2 and PCE contributing to 24.4, 17.7 and 12.2% of the total variance). Eigenvalues and the total percentage of variance of each PC axis for forewing shape are given in Supplementary table S1. PCA analysis plots of the studied lineages/species showed broad overlapping of wing shape among the three species, but with statistically significant differences. There is a discrimination of A. banksae and A. smithi (fig. 5) along the first axis, while A. eadyi slightly separated from the other two lineages along PC1. In the morphospace defined by PC2 and PC3, A. banksae separated slightly, with the populations from Israel having the most positive scores along PC3. Shape differences along the first three axes are presented in fig. 6.
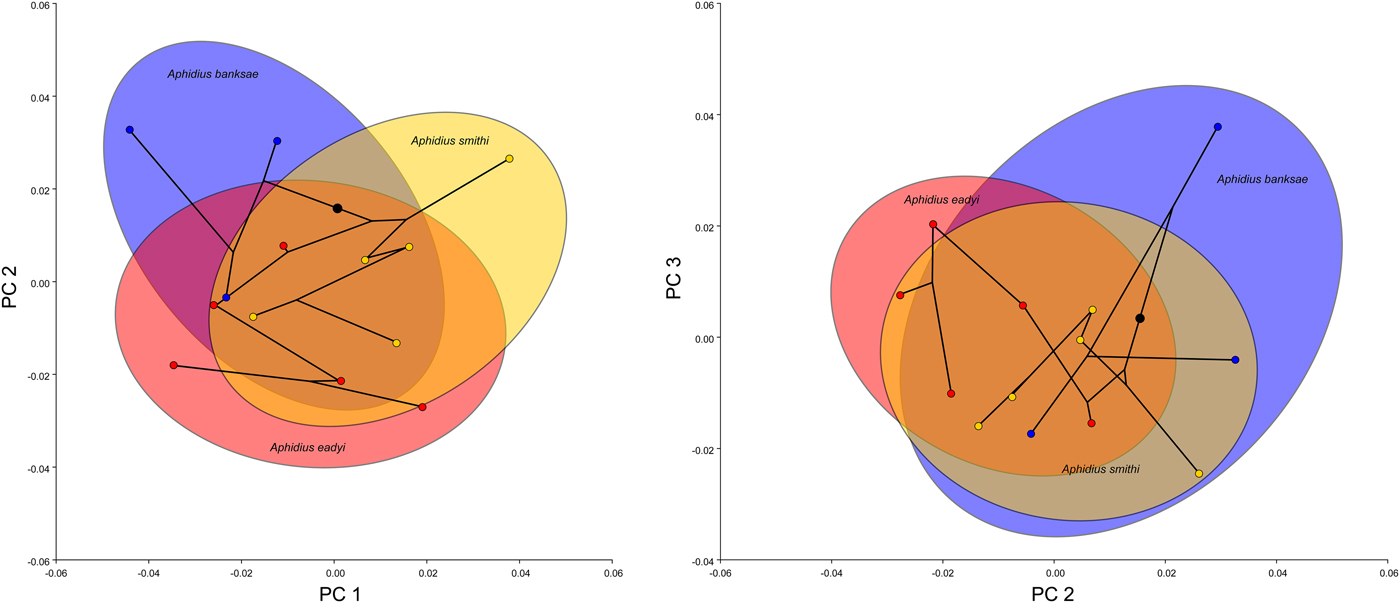
Fig. 5. Bivariate plots of mean PC-scores for the first three PC axes of forewing shape along with the superimposed phylogeny. The ellipses are sized so as to comprise 90% of the observations referring to the three phylogenetic lineages/species. The colour code is the same as in fig. 4: red – Aphidius eadyi, yellow – A. smithi, blue – A. banksae.

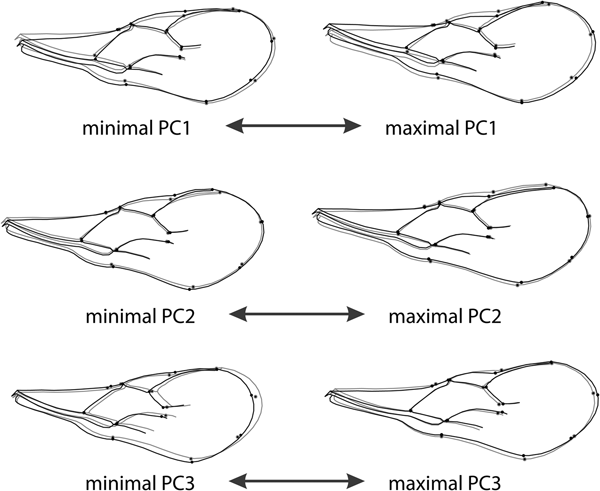
Fig. 6. Forewing shape changes associated with the first three PCs. The black outline represents the shape at the maximal positive and negative scores of each axis compared with the mean shape for the sample (grey).
Shape changes along PC1 are related to changes of the proximal part of the wing described by landmarks 2, 5, 6, 7, 8 and 9 and shape of the stigma (landmarks 2, 3, 4 and 5). The PC1 axis separated relatively shorter and wider wings with a wider proximal part, more robust stigma and longer radial vein, from wings with a narrower proximal part of the wing, narrower stigma and relatively shorter radial vein. The PC2 axis separated relatively wider wings with a concave anterior margin defined by the stigma and radial nerve (landmarks 1, 2, 3 and 4) from wings with a more or less flattened anterior margin of the forewing, such as those found in populations of A. banksae from Serbia and Israel. The PC3 axis separated relatively shorter and wider forewings with a shorter radial vein relative to the stigma (negative end of the PC3 axis) from more elongated wings with a longer radial vein (fig. 6).
The species average shape and visualization of shape differences between species are presented in fig. 7. The Procrustes distances are as follows: 0.022 between A. eadyi and A. smithi, 0.026 between A. eadyi and A. banksae, and 0.030 between A. smithi and A. banksae. All distances were statistically significant (P < 0.0001 in all comparisons).
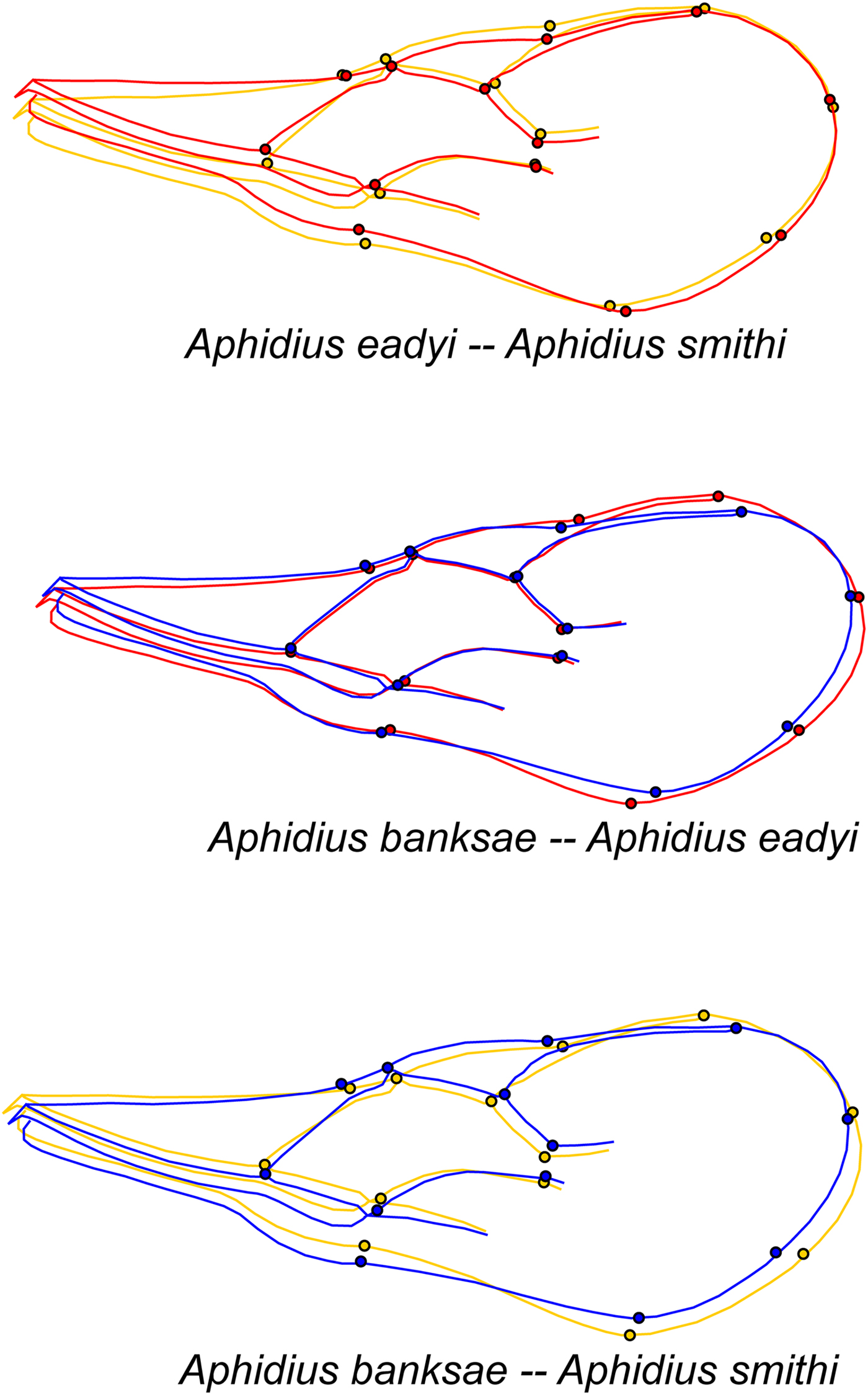
Fig. 7. Illustration of wing shape differences between the three analysed Aphidius species. Shape changes are shown as the difference between the average shape of the compared species. The colour code is the same as in fig. 4: red – A. eadyi, yellow – A. smithi, blue – A. banksae. All changes are exaggerated three times.
Discriminant function analysis showed that based on the forewing shape, a large proportion (>75%) of individual forewings in the confusion matrix is assigned to the correct species (table 3).
Table 3. Assignment of individual forewings to species as misclassified/number of specimens investigated.

Values from the Discrimination function analysis and those obtained through cross-validation were given below and to the right of the diagonal, respectively. All species combinations are above 75% of correct classification.
Morphology
After examining morphological characters commonly used in Aphidius taxonomy (Starý et al., Reference Starý, González and Hall1980), we found that the most important ones for separating species of the A. eadyi group are the number and shape of costulae on the anterolateral area of the petiole, shape of the central areola on the propodeum, and shape and venation of the forewings.
Redescription of A. banksae Kittel
(fig. 8)

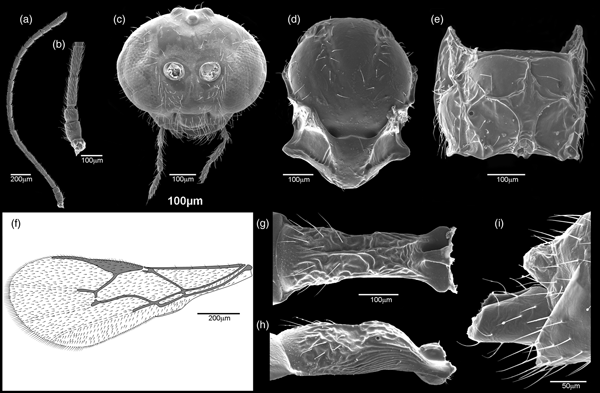
Fig. 8. Aphidius banksae, female. (a) Antenna, (b) first antennal segments, (c) frontal view of head, (d) dorsal aspect of mesonotum, (f) forewing, (e) propodeum, (g) dorsal aspect of petiole, (h) anterolateral area of petiole, (i) last genital segment and ovipositor sheath.
Material examined
ex A. pisum – Belgium: 1♀, Sint-Truiden (PCF), Lotus corniculatus, 14.x.2014. (AA); 1♀, Sint-Truiden (PCF) Trifolium repens, 07.x.2014. (AA); 1♀, Sint-Truiden (PCF), Trifolium sp., 16.ix.2014. (AA). Greece: 11♀ 7♂, Athens-Attica, Lathyrus cicera, 09.iv.1996. (NGK); 12♀ 6♂, Athens-Attica, Lathyrus clymenum, 23.iv.1997. (NGK); 4♀, Athens-Attica, Lathyrus ochrus, 23.iv.1997. (NGK); 6♀ 6♂, Athens-Attica, Lathyrus sativus, 04.iv.1996. (NGK); 14♀ 3♂, Athens-Attica, Lens culinaris, 09.iv.1996. (NGK); 1♀, Neohorion-Ilia, Medicago disciformis, 27.iv.1997. (NGK); 10♀ 7♂, Kopais-Voiotia, Medicago sativa, 09.v.1996. (NGK); 3♀1♂, Amalias-Ilia, Melilotus officinalis, 12.v.1997. (NGK); 1♀, Athens-Attica, Melilotus sulcata, 23.iv.1997. (NGK); 2♀, Amalias-Ilia, Trifolium nigrescens, 12.v.1997. (NGK); 1♀, Athens-Attica, Vicia narbonensis, 23.iv.1997. (NGK); 1♀ 1♂, Athens-Attica, Vicia pannonica, 01.iv.1997. (NGK); 3♀, Athens-Attica, Vicia sativa ssp. amphicarpa, 04.iv.1996. (NGK); 13♀ 4♂, Athens-Attica, V. sativa 04–26.iv.1996. (NGK); 3♀ 1♂, Athens-Attica, Vicia villosa, 23.iv.1997. (NGK). Israel: 48♀ 90♂, Beirut Sheian (Insectary Riverside-USA), M. sativa, 1979 (DG); 1♀ 14♂, Afigim (Insectary Riverside), M. sativa (DG). Montenegro: 4♀ 3♂, Tivat Vicia cracca, 25.v.2011. (AP); 1♂, Tivat, V. cracca, 25.v.2011. (VŽ). Serbia: 2♀ 2♂, Zemun, L. corniculatus, 12.v.2011. (AP); 21♀ 10♂, Živkovac, M. sativa, 3.vi.2012. (MJ); 18♀ 2♂, Reka, M. sativa, 6.vi.2012. (MJ); 2♀, Pančevački rit, M. sativa, 7.vi.2010. (MJ); 1♂, Umčari, M. sativa, 8.vi.2012. (MJ); 1♂, Malo Orašje, M. sativa, 8.vi.2012. (MJ). Slovenia: 1♀, Strujan, M. sativa, 20.xi.2008. (KK); 1♀, Strujan, M. sativa, 20.xi.2008. (KK); 1♀, Nova Gorica, M. sativa, 30.ix.2008. (KK).
Diagnosis
Aphidius banksae belongs to the A. eadyi group by virtue of sharing the group's host range pattern and wing venation. It differs from A. eadyi in having a longer R1 (fig. 8f) (stigma length/R1 length = 1.1–1.35 in A. banksae vs. 1.5–2.2 in A. eadyi) and a propodeum with a pentagonal areola that is wide anteriorly (fig. 8e), while it is narrow in A. eadyi. Aphidius banksae differs from A. smithi in having 7–14 irregularly curved costulae on the anterolateral area of the petiole (fig. 8h), while there are 4–6 almost straight costulae in A. smithi.
Description
Female: Head (fig. 8c) wider than mesosoma at tegulae (head width/mesoscutum width = 1.31–1.44). Frons, vertex and occipital area with dense setae. Face moderately setose (fig. 8c). Tentorial index 0.45–0.55. Malar space equal to 0.25–0.35 of longitudinal eye diameter. Eyes oval, converging towards clypeus. Clypeus rounded, with 7–13 long setae. Antennae 19-segmented, very rarely 20-segmented, uniformly filiform (fig. 8a), with semi-erected and adpressed setae, which are 1/4 shorter than segment diameter. Scape and pedicel subglobular. Flagellomere 1 (F1), 3.00–4.00 times as long as its maximum width (fig. 8b). F2 3.00–4.30 times as long as its maximum width. F1 somewhat shorter than or subequal to F2 (F1l/F2l = 0.85–0.93). F1 and F2 without and with three longitudinal placodes, respectively. Maxillary palps with four palpomeres. Labial palps with three palpomeres.
Mesosoma: Mesonotum with notaulices in the ascendant portion of its anterolateral area, erased dorsally and outlined by two rows of long sparse setae (fig. 8d). Scutellum with 5–6 short setae, mostly in lateral parts. Forewing (fig. 8f) stigma moderately elongated, 3.00–3.55 times as long as its width, ratio between stigma length and R1 1.10–1.35. Propodeum (fig. 8e) areolated with a pentagonal central areola, wide anteriorly and narrow posteriorly. Upper areolae with 2–3 long setae laterally and lower areolae with 2–4 setae. Hind femur and tibia with semi-erected sparse setae.
Metasoma: Petiole almost parallel-sided (fig. 8g), 3.10–3.70 times as long as its width at spiracles, anterolateral area with 7–14 irregularly curved costulae (fig. 8h). Dorsal surface of the petiole with fine rugosities, moderately prominent mediodorsal carina and 15 long semi-erected lateromedial setae on its lower half (fig. 8g).
Genitalia: Ovipositor sheath (fig. 8i) slightly concave at the dorsal margin.
Coloration: Head brown with black eyes, face and genae yellow to light-brown, mouthparts yellow. Scapus and pedicel light-brown to yellowish, annellus yellow, remaining parts of flagellum uniformly brown except for a narrow yellow ring at the base of F1. Pronotum yellow. Mesonotum light-brown to brown with a light-brown metapleuron. Legs yellow with dark apices. Wings hyaline. Metasoma (including petiole) light-brown to yellowish with a dark-brown ovipositor sheath. According to the original description, the colour may vary seasonally (Chen et al., Reference Chen, González and Luhman1990).
Body length: ~3 mm.
Male: Antennae 20–21-segmented. Generally darker than the female. Scapus and pedicel yellow to light-brown. Face and mouthparts light-brown. Pronotum light-brown. Legs yellow with dark apices. Remaining body parts brown.
Hosts
Acyrthosiphon pisum.
Distribution
On the basis of our research, it can be asserted that A. banksae is distributed in several European countries (Belgium, France, Greece, Montenegro, Serbia, Slovenia, UK), as well as in the Middle East (Israel). According to Chen et al. (Reference Chen, González and Luhman1990), A. banksae has also been recorded in Turkey. We assume wider distribution of A. banksae throughout Europe in association with A. pisum.
Discussion
Any meaningful research in biology is based on reliable taxonomy (Rosen, Reference Rosen1986; Moraes, Reference Moraes1987; Schlick-Steiner et al., Reference Schlick-Steiner, Steiner, Seifert, Stauffer, Christian and Crozier2010; Mehle & Trdan, Reference Mehle and Trdan2012; Dorchin et al., Reference Dorchin, Joy, Hilke, Wise and Abrahamson2015). Accordingly, accurate identification of natural enemies is an essential element for the success of biological control programmes (Rosen, Reference Rosen1986; Moraes, Reference Moraes1987). Nowadays, integrative taxonomy is the only reliable tool because it goes beyond naming of species and gives priority to species delineation and processes underlying it (Dayrat, Reference Dayrat2005; Schlick-Steiner et al., Reference Schlick-Steiner, Steiner, Seifert, Stauffer, Christian and Crozier2010). Our integrative taxonomic research on the A. eadyi species group resulted in clear separation of three species, viz., A. smithi, A. eadyi and A. banksae. This separation is based on mtDNA COI sequences, morphometrics and morphology.
Mean genetic distances between all three species (ranging from 5 to 7.4%) are above the common rate for between-species divergence in the genus Aphidius (Kos et al., Reference Kos, Petrović, Starý, Kavallieratos, Ivanović, Toševski and Tomanović2011; Tomanović et al., Reference Tomanović, Petrović, Mitrović, Kavallieratos, Starý, Rakhshani, Rakhshanipour, Popović, Shukshuk and Ivanović2014). Apart from the high intraspecific genetic variation recorded in A. smithi and to a lesser extent in A. eadyi, all phylogenetic trees were the same, with haplotypes of all three species clustered together. Although all differences of wing shape in all three species were statistically significant, there was a broad overlapping of wing shape among the three species. The most divergent wing shape was recorded in A. banksae, which is congruent with molecular analysis. Similar results were obtained by Tomanović et al. (Reference Tomanović, Petrović, Mitrović, Kavallieratos, Starý, Rakhshani, Rakhshanipour, Popović, Shukshuk and Ivanović2014) for the A. colemani species group.
The highest intraspecific genetic divergence was recorded in A. smithi, where eight haplotypes differ from each other in the range of 0.2–4.3% (mean 2.1%). Given that some of these differences exceed the intraspecific genetic variation in Aphidius (Kos et al., Reference Kos, Petrović, Starý, Kavallieratos, Ivanović, Toševski and Tomanović2011; Tomanović et al., Reference Tomanović, Petrović, Mitrović, Kavallieratos, Starý, Rakhshani, Rakhshanipour, Popović, Shukshuk and Ivanović2014; Derocles et al., Reference Derocles, Plantegenest, Rasplus, Marie, Evans, Lunt and Le Ralec2016), it follows that several of them could possibly represent some cryptic species. This is supported with the results of PTP and ABGD, which also suggested the presence of independent lineages. However, additional research efforts are needed to confirm this hypothesis, because at this point, we were unable to find any morphological, ecological or biological characters to distinguish between these entities. The most divergent haplotypes were Asmit1 from Turkey and Asmit2 from Brazil, which differed by more than 2% from all other haplotypes and also from each other. Such high genetic differences within A. smithi cannot be explained either by influence of the host (all originated from the same host) or by geography. The genetically closest relatives (Asmit3, Asmit4 and Asmit5) were from Afghanistan, Uzbekistan and Spain. One possible explanation of such genetic diversity of A. smithi is that it originates in the native range of the species (India). Circumstantial evidence can be found in comparison of Asmit8 and Asmit6, which differed by 1.5%. Asmit8 was initially collected from India and reared for mass release in an insectarium in Riverside, while Asmit6 was collected in Lakeview (CA, USA) as a sample recovered after initial establishment. During the 1970s, there were several long-term biocontrol projects targeting the management of A. pisum in the USA that resulted in certain studies about the biology and ecology of A. smithi (see Starý, Reference Starý1974; Angalet & Fuester, Reference Angalet and Fuester1977). However, there is a very big gap in knowledge about the current status and distribution of A. smithi in North America. Angalet & Fuester (Reference Angalet and Fuester1977) reported that A. smithi was displaced by A. ervi all over the USA and became almost extinct in North America (McBrien & Mackauer, Reference McBrien and Mackauer1990). Potentially, it still exists in low-density populations with no useful agricultural effect on the pest (Wylie et al., Reference Wylie, Matheson, Uddin and Holliday2005). In the light of results of the above studies and our own findings, it is clear that a detailed survey is needed to ascertain the current existence and status of A. smithi in North America. We also determined the existence of A. smithi in Europe (Asmit3 from Spain), which is in agreement with published data (Pennacchio, Reference Pennacchio1989; Rasplus et al., Reference Rasplus, Villemant, Paiva, Delvare and Roques2010; Yu et al., Reference Yu, van Achterberg and Horstmann2012; van Achterberg, Reference Van Achterberg2013). Although the presence of A. smithi in Europe is not open to question, its origin and current distribution certainly are obscure, with just a few samples available. Starý (Reference Starý1974) asserted that A. smithi was introduced and established in hot and dry areas of Europe prior to the official unsuccessful releases in Central Europe. Unfortunately, we were unable to make any conclusion as to the origin of A. smithi in Europe since we analysed only one available European population of this species. Based on the mtCOI sequences, it can be concluded that the Spanish population of A. smithi (Asmit3) is closely related to the populations from Afghanistan and Uzbekistan. A critical review of all relevant literature (summarized in Yu et al., Reference Yu, van Achterberg and Horstmann2012) indicates that the distribution of A. smithi is largely overestimated. In view of its climatic requirements (Campbell & Mackauer, Reference Campbell and Mackauer1973; Starý, Reference Starý1974) and the Oriental origin of A. smithi, we conclude that it is distributed in Mediterranean Europe. This is also supported by the fact that the only relevant findings are from Spain (herein), Italy (Pennacchio, Reference Pennacchio1989) and Greece (Kavallieratos et al., Reference Kavallieratos, Tomanović, Starý, Athanassiou, Sarlis, Petrović, Niketić and Anagnou Veroniki2004), whereas records from Bulgaria (Atanassova, Reference Atanassova1997) and Turkey (Akar & Çetin Erdoğan, Reference Akar and Çetin Erdoğan2017) should be taken with caution. We also analysed A. smithi from Turkey (Asmit1), but our specimens were collected in central Anatolia, so they cannot be treated as Europe. All other records of A. smithi should be re-evaluated in light of the present results, especially those from the studies of Rasplus et al. (Reference Rasplus, Villemant, Paiva, Delvare and Roques2010) and Yu et al. (Reference Yu, van Achterberg and Horstmann2012), in both of which more than 25 countries are listed.
The analysed specimens of A. eadyi were assigned to six different haplotypes with mean intraspecific genetic divergence of 1.5%. This intraspecific genetic variability is considered very high when compared with the overall variability (≤0.5%) of the recently analysed A. colemani species group (Tomanović et al., Reference Tomanović, Petrović, Mitrović, Kavallieratos, Starý, Rakhshani, Rakhshanipour, Popović, Shukshuk and Ivanović2014) and A. urticae s. str. group (Jamhour et al., Reference Jamhour, Mitrović, Petrović, Starý and Tomanović2016). The bulk of the detected divergence is caused by one haplotype (Aeady6) from the Czech Republic, which differs from all others by ≥2.3%. This specific haplotype is recognized as an independent entity by both PTP and ABGD. It is worth mentioning that the haplotypes Aeady3 and Aeady4 also originated from the Czech Republic, but they are genetically closer to haplotypes from other parts of Europe (Aeady1 and Aeady2) and Iran (Aeady5) than to Aeady6. Our results constitute molecular confirmation of the West Palaearctic distribution of A. eadyi postulated by Starý et al. (Reference Starý, González and Hall1980). A. eadyi was used as a BCA against A. pisum in New Zealand, where it had a fate similar to that of A. smithi in North America, i.e., it was displaced by A. ervi (Cameron & Walker, Reference Cameron and Walker1989). There are no recent data about A. eadyi in the introduced areas (Burundi and New Zealand), and thus its current status is unknown.
The most interesting finding of our study is the identification of A. banksae as a relatively common and widely distributed parasitoid of the pea aphid in the Western Palaearctic. After analysing the mtCOI sequences as well as the forewing shape, it became evident that A. banksae is unambiguously a distinct species, one that was previously treated as A. urticae (Kavallieratos et al., Reference Kavallieratos, Tomanović, Starý, Athanassiou, Sarlis, Petrović, Niketić and Anagnou Veroniki2004; Alhmedi et al., Reference Alhmedi, Haubruge and Francis2009; Žikić et al., Reference Žikić, Ilić-Milošević, Stanković, Petrović, Petrović-Obradović, Kavallieratos, Starý and Tomanović2012; Derocles et al., Reference Derocles, Plantegenest, Rasplus, Marie, Evans, Lunt and Le Ralec2016) or as A. eadyi (Elias et al., Reference Elias, Fontaine and van Veen2013). Since we found some morphological differences in relation to the original description of A. banksae (Chen et al., Reference Chen, González and Luhman1990), we are re-describing this species on the basis of a series of specimens collected throughout Europe; and specimens from a population reared at the University of California, Riverside, which originated from the type locality in Israel (Beirut Sheian). Unfortunately, the holotype and paratypes from the Smithsonian National Museum of Natural History (NMNH) were not available to us for re-examination. The observed differences were mainly in a range of characters wider than previously recorded. Also, the original diagnosis was mainly based on colour, which can be highly variable, whereas we used numerical characters commonly employed in Aphidius taxonomy (Starý, Reference Starý1974; Pennacchio, Reference Pennacchio1989; Tomanović et al., Reference Tomanović, Petrović, Mitrović, Kavallieratos, Starý, Rakhshani, Rakhshanipour, Popović, Shukshuk and Ivanović2014). Aphidius banksae is characterized by the highest number of identified haplotypes, but has the lowest mean intraspecific genetic variation within the A. eadyi species group. Moreover, no evident association with a specific geographic region was determined for A. banksae haplotypes. Chen et al. (Reference Chen, González and Luhman1990) stated that A. banksae is distributed in Turkey and Israel and thus is allopatric in relation to A. eadyi, while results of its introduction in the USA are unknown. On the other hand, our results indicate a much broader distribution (from the UK to Israel) for A. banksae and its sympatry with A. eadyi. Apart from their almost identical geographic distribution, both species parasitize A. pisum attacking several plants of the family Fabaceae (see table 1 and Starý et al., Reference Starý, González and Hall1980).
Although sympatric speciation is common in the Aphidiinae, it cannot be the case with the A. eadyi group, because speciation in it is mostly driven by parasitoid specialization to different aphid host lineages (Tremblay & Pennacchio, Reference Tremblay, Pennacchio and Gupta1988; Kos et al., Reference Kos, Petrović, Starý, Kavallieratos, Ivanović, Toševski and Tomanović2011; Mitrovski-Bogdanović et al., Reference Mitrovski-Bogdanović, Petrović, Mitrović, Ivanović, Žikić, Starý, Vorburger and Tomanović2013; Tomanović et al., Reference Tomanović, Petrović, Mitrović, Kavallieratos, Starý, Rakhshani, Rakhshanipour, Popović, Shukshuk and Ivanović2014; Jamhour et al., Reference Jamhour, Mitrović, Petrović, Starý and Tomanović2016). Aside from the fact that the pea aphid is a complex of host-specialized races and species (Peccoud et al., Reference Peccoud, Ollivier, Plantegenest and Simon2009a), with one of the fastest rates of evolutionary diversification ever recorded (Peccoud et al., Reference Peccoud, Simon, McLaughlin and Moran2009b), our evidence indicates that there is no host-related specialization of the A. eadyi species group, since in several cases, A. banksae and A. eadyi were reared from the same aphid colony (Aeady1 and Abank1; and Abank3, Abank8 and Abank11). Also, it was common to collect A. banksae and A. eadyi in the same sample where A. banksae was erroneously identified as A. urticae or as a light form of A. eadyi. Based on the obvious sympatry and their independent evolution over a relatively long period of time (genetic divergence of 7.4%), it would appear that A. banksae and A. eadyi acquired the pea aphid independently. Aphidius banksae is genetically more closely related to A. smithi than to A. eadyi, and it can thus be suspected of having originated in the Middle East, from where it was described (Chen et al., Reference Chen, González and Luhman1990). It can be assumed that the Middle East is the centre of diversity for the A. eadyi species group, since it is most likely that all three species naturally cohabit there.
Programmes for biological control of A. pisum after the 1980s were concentrated on A. ervi because it was proved to be a better competitor in comparison with A. eadyi and A. smithi (Angalet & Fuester, Reference Angalet and Fuester1977; Cameron & Walker, Reference Cameron and Walker1989; McBrien & Mackauer, Reference McBrien and Mackauer1990). On the other hand, the existence of symbiont-conferred resistance to parasitoids in the pea aphid (Oliver et al., Reference Oliver, Russell, Moran and Hunter2003) has the potential to compromise the effectiveness of biological control (Vorburger, Reference Vorburger2018). Numerous studies have confirmed that defensive symbionts of the pea aphid can protect that pest from A. ervi (Vorburger, Reference Vorburger2018), but only one showed symbiont-conferred resistance to A. eadyi (Ferrari et al., Reference Ferrari, Darby, Daniell, Godfray and Douglas2004).
Our results shed light on the taxonomic status of species belonging to the A. eadyi group, as well as on genetic diversity of the three analysed species, providing information which could be very useful in future biological control strategies. One of the recommendations for future successful biocontrol strategies is to overcome the symbiont-conferred resistance of aphid pests by maintaining high genetic diversity of stock parasitoids (Vorburger, Reference Vorburger2018). With relatively high intraspecific genetic diversity, A. banksae, A. eadyi and A. smithi are good candidates for such an approach. Our study raises issues regarding the current distribution of BCAs belonging to the A. eadyi species group in the areas of introduction. These questions can be answered by conducting an additional exhaustive survey of pea aphid parasitoids. Also, the origin of A. banksae and to some extent of that A. eadyi are still open issues that could be resolved by performing a phylogeographic analysis of the overall species distribution.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S000748531800055X.
Acknowledgements
This study was supported by Grant III43001 from the Ministry of Education, Science, and Technological Development of the Republic of Serbia. The participation of P. Starý was made possible by institutional support.