Introduction
Phytophagous insects colonise a wide range of plant host species by two strategies. The first is ‘specialisation’, whereupon only a restricted range of host plants is colonized, and this limits the need for adaptability at an individual level. The second is for individuals to be adaptable as ‘generalists’, able to colonise many host plant species. Specialists benefit from expending fewer resources in countering a range of plant defence compounds (Despres et al., Reference Despres, David and Gallet2007). Generalists are capable of varying their physiology, creating greater ecological plasticity and increasing the likelihood of finding suitable host plants all year round (Despres et al., Reference Despres, David and Gallet2007). It is also possible that a mixture of specialists and generalists could be represented by different genotypes within a species.
For many insects, studying the level of individual adaptability is complicated by genetic variation such that no two individuals are the same and genetic background is a factor which cannot be easily eliminated in ecological studies. This complication is partly overcome by studying aphids which generate diversity through sex for one generation per year, but increase summer populations through many generations of clonal reproduction where genetic variation is constrained to spontaneous mutation (Loxdale & Lushai, Reference Loxdale and Lushai2003). Clones best adapted to their environment will become abundant in both space and time, and their quantity is to some extent a measure of their success.
Many aphid species are specialists, colonising only one plant species. For example, Amphorophora idaei Börner colonises raspberry (Rubus idaeus L.) and A. rubi (Kalt.) colonises blackberry (Rubus fruticosus L.) (Blackman & Eastop, Reference Blackman and Eastop1984). Other aphid species appear to colonise many crop plants, but careful study finds that some of these species exhibit varying degrees of host plant specialism at the clone level (Blackman, Reference Blackman, Campbell and Eikenbary1990). Specialized clones have been found in the greenbug Schizaphis graminum (Rondani) (Shufran et al., Reference Shufran, Burd, Anstead and Lushai2000), grain aphid Sitobion avenae (Fabricius) (Caillaud et al., Reference Caillaud, Dedryver, Di Pietro, Simon, Fima and Chaubet1995; De Barro et al., Reference De Barro, Sherratt, David and Maclean1995), pea aphid Acyrthosiphon pisum (Harris) (Via, Reference Via1991) and cabbage aphid Brevicoryne brassicae (L.) (Ruiz-Montoya et al., Reference Ruiz-Montoya, Núñez-Farfan and Vargas2003). The peach-potato aphid, Myzus persicae (Sulzer), colonises some 300 plant species spread over 72 families worldwide (Gladders & Peters, Reference Gladders and Peters1986). How this adaptability is achieved at the individual level is far from clear. In Germany, Weber (Reference Weber1985) studied over 1000 clonal lineages of M. persicae collected mainly from potato and sugar beet fields. Some clonal lineages reproduced better on the original source plant than on an alternative species; and this performance was stable over several generations, suggesting some clones were better adapted to particular hosts than others. Other clones performed equally well on both hosts, providing evidence for generalism. Edwards (Reference Edwards2001) also found significant variation in the mean relative growth rates between M. persicae clonal lineages grown on lupin but no differences on chickpea, lentil, pea or faba bean. Weber (Reference Weber1985) and Edwards (Reference Edwards2001) did not characterise their lineages genetically so did not know if variability was associated with inter- or intra-clonal differences. Vorburger et al. (Reference Vorburger, Lancaster and Sunnucks2003a,b) genotyped Australian M. persicae clonal lineages and compared the reproductive potential of anholocyclic or androcyclic clones (asexual all year round) with those that were holocyclic (returning to peach for a sexual generation). Vorburger et al. (Reference Vorburger, Sunnucks and Ward2003b) hypothesized that widespread asexual clones would be more generalist than sexual clones, as they have been subjected to greater plant selection on secondary hosts (Vorburger et al., Reference Vorburger, Sunnucks and Ward2003b). While differences between clones were found, there was no correlation between lifecycle and their performance on different host plants.
There is clear evidence for one M. persicae specialist, a race associated with tobacco. This race can be distinguished from aphids originating from other host plants, genetically, morphologically and at the population level (Blackman, Reference Blackman1987; Blackman & Spence, Reference Blackman and Spence1992; Margaritopoulos et al., Reference Margaritopoulos, Mamuris and Tsitsipis1998, Reference Margaritopoulos, Malarky, Tsitsipis and Blackman2007; Nikolakakis et al., Reference Nikolakakis, Margaritopoulos and Tsitsipis2003; Blackman et al., Reference Blackman, Malarky, Margaritopoulos and Tsitsipis2007).
In field studies of Scottish M. persicae clones, there was little evidence of clone host-plant association, on either potato or brassicas, in 1995 and 2001 (Fenton et al., Reference Fenton, Woodford and Malloch1998, Reference Fenton, Malloch, Woodford, Foster, Anstead, Denholm, King and Pickup2005). However, in later years, evidence of host specialized and localized clones has been found, and the later populations also included successful insecticide resistant clones (Kasprowicz et al., Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a,Reference Kasprowicz, Malloch, Pickup and Fentonb).
The current study investigates the plant-associated reproductive performance of lineages of clones that comprise the well-characterized Scottish M. persicae population. The relative numerical success of these clones has been studied over a ten-year period. Three clones have made up most of the individuals in the population for at least the past 20–30 years and they are considered successful. Other clones have varying levels of success with some being very successful, in short periods of one or two years, but disappearing after that. Yet others are found for a longer period of time but never in great numbers (Fenton et al., Reference Fenton, Malloch, Woodford, Foster, Anstead, Denholm, King and Pickup2005; Kasprowicz et al., Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a, Reference Kasprowicz, Malloch, Pickup and Fentonb). In this paper, we addressed the following questions: Do clones differ in reproductive success on different host plants that would indicate specialists and generalists? Is reproductive success on common agricultural host plants correlated with field success? How is clone performance affected by temperature? Is there any evidence of reproductive costs associated with insecticide resistance?
Materials and methods
Clonal lineages
The plant-associated reproductive performances of 18 clonal lineages of M. persicae on different host plants were investigated using experimental procedures adapted from Vorburger et al. (Reference Vorburger, Sunnucks and Ward2003b). For convenience, the clones have been divided into the following categories: long-term insecticide sensitive, new insecticide resistant invader, long-term insecticide resistant, host specialized or localized and numerically abundant or rare (table 1). Eighteen lineages were used (table 1; for details, see Kasprowicz et al., Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a,b). Four of these lineages represented two clone duplicates (clone H=K5 and K10; clone J=159 and Mp1S). For all lineages, two independent trials were used (either ten pots or 20 pots for each lineage). To check for contamination, the colonies were genotyped with microsatellite markers at the beginning and end of the project, and none was found.
Table 1. Laboratory maintained clonal lineages of M. persicae.

Column 1 provides the letter for the multilocus genotype and the parentheses lineage names for lineages with duplicates. For details of these clones see Kasprowicz et al. (Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a,Reference Kasprowicz, Malloch, Pickup and Fentonb). Column 2 indicates the year the lineage was first collected in the field and column 3 the category of clone. Columns 4–7 denote the insecticide resistance characteristics of that clone. The esterase is expressed as resistance (R levels), with S being sensitive and R3 being the most resistant. Esterase confers resistance to insecticides such as organophosphates. MACE=modified acetylcholinesterase (resistance to dimethyl carbamates); kdr and skdr knock down and super knock down (resistance to pyrethroids).
Plant material
Seedlings of four plant species were used as hosts: (i) downy ground-cherry, Physalis floridana; (ii) radish, Raphanus sativus L., var. Long White Icicle; (iii) spring oilseed rape Brassica napus (OSR) var. Mascot; and (iv) potato Solanum tuberosum L., var. Desiree. Radish was used as a point of reference to Vorburger et al. (Reference Vorburger, Sunnucks and Ward2003b). OSR and potato are common M. persicae hosts in Scotland. P. floridana is often used as a virus indicator plant for work with M. persicae. Experiments followed the procedure of Vorburger et al. (Reference Vorburger, Sunnucks and Ward2003b) with two exceptions: (i) temperature was held constant at 18°C rather than at 20°C; (ii) a single nymph of a known age was applied to the plant instead of placing reproducing females on the plant overnight and removing all but one nymph the next day. For M. persicae, 18–20°C is close to the optimum temperature for maximum parthenogenetic reproduction (Liu & Meng, Reference Liu and Meng1999), and this would correspond to its temperate origins and sexual reproduction associated with peach trees.
Growth measurements
From independent colonies of each lineage adapted to each host plant for at least ten generations, 20 one-day-old nymphs were placed singly onto 20 individual seedlings. The seedlings and pots were covered with a clear Perspex® tube (8 cm external diameter×7 cm internal diameter×16 cm length; Stockline Plastics, UK; referred to as pots). To retain aphids while allowing air-flow, each tube was capped with a thin mesh (mesh size 200 μm; John Lewis, UK) held in place with a strong rubber band. The pots were then placed in a controlled environment room at 18±2°C on a 16:8 h light:dark cycle for 15 days. On day 15, the experiment was terminated by cutting each plant stem and placing it, and the aphid colony, into screw-topped tubs (6.5 cm×7.5 cm; VWR, UK). These were then frozen and stored at −20°C until the individuals in the colony could be counted by carefully disassembling each plant. Each treatment was repeated as above, except potato, where the second trial was of ten pots. Pots that had no colony, or no founding aphid, when the treatment ended were not included in the analysis.
The reproductive performance of lineages at 14°C was also investigated but only on OSR. This temperature is closer to the average summer temperature for Scotland but is still warm enough to enable development in 15 days. The experiments were carried out as above, except two trials of ten pots were used.
A small experiment was set up in a controlled environment growth chamber (MLR-350, Sanyo, UK) to replicate, as closely as possible, the conditions used by Vorburger et al. (Reference Vorburger, Sunnucks and Ward2003b). The light:dark cycle was 16:8 h and the temperature was a constant 20°C. Lineages of two clones, G and K, were used. Eight one-day-old nymphs of each lineage were placed individually onto four OSR or four radish seedlings and left for 15 days.
Statistical analysis
The effects of host plant, clonal lineage and their interactions on the average colony count (abbreviated to count) after 15 days at 18°C were analysed using untransformed values in an unbalanced general ANOVA (Genstat, 8th Edition, VSN International). The analysis was adjusted for any effect of the replication. The data were analysed as a whole, then for the effect of clonal lineage and its interaction with the host plant for each host. For the comparison between the average count of two clonal lineages on two host plants at 18°C and 20°C, the effects of host plant, clonal lineage and temperature on count, and their interactions, were analysed using a general ANOVA. The effects of changing the temperature, from 18°C to 14°C, on count and clonal lineage were also investigated using untransformed data and an unbalanced general ANOVA.
All comparisons between pairs of clonal lineages in all experiments were investigated using two-sided t-tests to perform pair-wise comparisons of the differences of the means from each ANOVA. Pearson's correlation coefficients were calculated with the aid of Microsoft Excel.
Results and discussion
Studies to determine what makes a particular M. persicae clone successful have examined reproductive performance in association with insecticide resistance fitness costs, life history characteristics, biochemistry and cold tolerance (Weber, Reference Weber1985, Reference Weber1986; Foster et al., Reference Foster, Denholm and Devonshire2000, Reference Foster, Denholm, Thompson, Poppy and Powell2005; Edwards, Reference Edwards2001; Vorburger et al., Reference Vorburger, Sunnucks and Ward2003b; Vorburger, Reference Vorburger2004; Francis et al., Reference Francis, Vanhaelen and Haubruge2005). A comparison between these studies has been difficult due to different aspects of insect physiology, different plant hosts, different or unknown sets of clones, or those for which the long-term ecological success is unknown. In areas of aphid sexual reproduction, genetic diversity and variations in life history characteristics are further complications (Vorburger et al., Reference Vorburger, Sunnucks and Ward2003b). In this paper, we investigated the reproductive potential of lineages representing clones in the Scottish M. persicae population. These clones have an understood ecology, and sexual recombination is not a factor in their recent origins (Fenton et al., Reference Fenton, Woodford and Malloch1998, Reference Fenton, Malloch, Woodford, Foster, Anstead, Denholm, King and Pickup2005; Malloch et al., Reference Malloch, Highet, Kasprowicz, Pickup, Neilson and Fenton2006; Kasprowicz et al., Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a,Reference Kasprowicz, Malloch, Pickup and Fentonb).
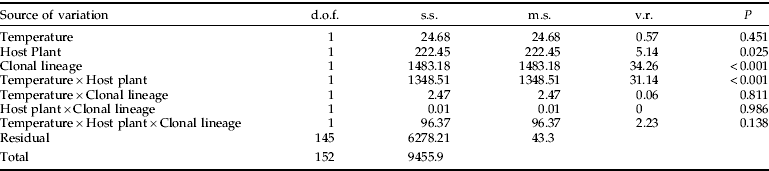
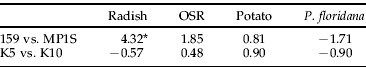
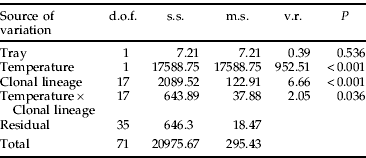
To determine whether differences in reproduction contribute to the relative field success of any clone, we used a procedure which measured the number of individuals in a colony produced from a one-day-old nymph after 15 days (Vorburger et al., Reference Vorburger, Sunnucks and Ward2003b). This measurement is similar to the 12-day measurements used by Weber (Reference Weber1985) and includes development time, fecundity and nymph mortality and, thus, is a good indicator of the intrinsic rate of increase of a lineage (Vorburger et al., Reference Vorburger, Sunnucks and Ward2003b). As our insectaries are shared facilities, we had to carry out large-scale experiments at 18°C, slightly cooler than the 20°C used by Vorburger et al. (Reference Vorburger, Sunnucks and Ward2003b). We established that there was no significant difference in measurements made at 18 and 20°C using lineages of clones G and K (table 2). There was a significant difference between the counts of the two clonal lineages at 20°C, with G having a larger count compared with K. This rank order is in agreement with the results at 18°C (see next section). There was a significant interaction between temperature and host plant (table 2). On radish at 20°C, the count for both lineages was significantly larger than on OSR; but, at 18°C, the count of both clones was significantly smaller on radish than OSR (fig. 1).

Fig. 1. A comparison of count (±SEM) after 15 days at 18°C and 20°C for clonal lineages G and K on radish and OSR (![]() , G (20°C);
, G (20°C); ![]() , G (18°C); □, K (20°C );
, G (18°C); □, K (20°C ); ![]() , K(18°C)).
, K(18°C)).
Table 2. An ANOVA of the count of two clone lineages on two host plants at 18°C and 20°C.
To directly compare our results to Vorburger (Reference Vorburger, Sunnucks and Ward2003b), a treatment was carried out at 20°C using lineages of clones G and K on OSR and radish. The 20°C treatments were compared using ANOVA. There was a significant difference between the counts of the two clonal lineages at 20°C, with G having a larger count than K and there was a significant interaction between temperature and host plant. d.o.f., degrees of freedom; s.s., sum of squares; m.s.. mean square; v.r., variance ratio; P, probability.
The count of M. persicae clonal lineages at 18°C
Having established that there was no significant difference between 18 and 20°C, the main experiment was continued at 18°C. Two independent trials for each combination of host plant and clonal lineage were completed, and there were no significant differences between any of the replicates (table 3). The count of M. persicae on the four host plants did differ significantly (fig. 2, table 3). The largest colony counts were produced on both OSR and radish (count ~39). Colonies on potato were generally smaller (count ~30), while colonies on P. floridana were the smallest (count ~18).

Fig. 2. The count after 15 days of 18 clonal lineages of M. persicae on four host plants at 18°C. The lineages with an identical multilocus microsatellite genotype are believed to derive clonally from the same stem mother. These are further differentiated by their unique lineage name in brackets. Note: clones I, D, N and E were not investigated on P. floridana. The order of the lineages was based on the OSR graph with the smallest value on the left and largest on the right. The error bars represent the standard error of the mean (□, OSR; ▪, Physalis; ![]() , Potato;
, Potato; ![]() , radish).
, radish).
Table 3. The results of significance tests from two independent trials of the average colony count after 15 days for each combination of host plant and clonal lineage.
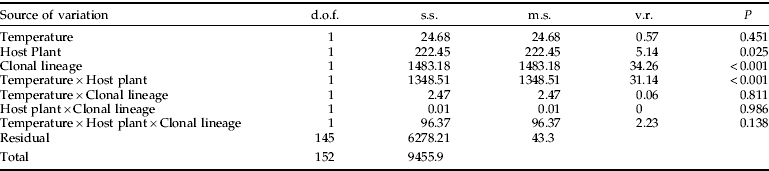
The independent trials of each lineage were not significantly different. The count of M. persicae (all lineages) on the four host plants differed significantly (tray). There was also a significant effect of clonal lineage, which suggested variation among the count of different lineages when on the same host.
* Denotes significant differences (P<0.05). For abbreviations see table 2.
There was also a significant effect of clonal lineage, which suggested variation among the counts of different lineages on the same host (fig. 2, table 3). On P. floridana, the largest count was produced by the M lineage (28), and the smallest was produced by the A lineage (9; fig. 2). On radish, lineage H (K5, 50) had the largest count, while lineage F had the smallest (19). On OSR, clone H (lineage K10, 52) was the largest count, and the smallest was lineage F (18). P9Bn and H (lineage K10) had the largest count on potato (both 37), and clone F produced the smallest (21).
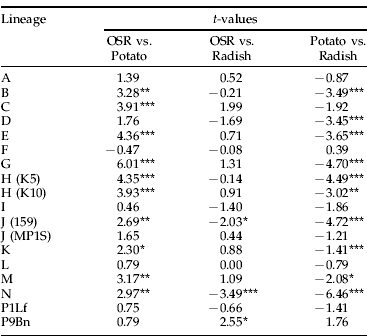
Duplicate lineages were used for two clones, J and H. There were no significant differences between the lineages within each clone apart from clone J on radish where lineage 159 had a significantly greater count than lineage MP1S (table 4).
Table 4. t-values from pairwise comparisons between the count of two clone J lineages (159 and MP1S) and two clone H lineages (K5 and K10).
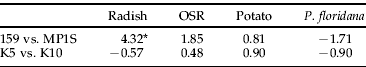
There were no significant differences between the lineages within each clone apart from clone J, where lineage 159 had a significantly greater count than lineage MP1S on radish.
* Denotes significant differences (P<0.05); df=63.
Compared to other host plants, P. floridana produced the smallest count for most of the lineages (fig. 2). There were some exceptions: clonal lineage F produced similar counts on all hosts, and lineages of B (t=1.62, df=35, P=0.109), G (t=1.16, df=35, P=0.252) and M (t=0.61, df=35, P=0.541) produced counts on potato that were not significantly different from their count on P. floridana. Oilseed rape and radish produced equally large colonies for most lineages. However, clonal lineages J (159) and N had significantly larger colonies on radish than on OSR, and clonal lineage P9Bn produced significantly larger colonies on OSR than on radish (table 5). All clonal lineages, apart from F, produced larger colonies on OSR than on potato, and ten of these were significantly larger (table 5). All but two clonal lineages produced larger colonies on radish than on potato, and ten were significantly larger. P9Bn produced larger colonies on potato than on radish, as did clonal lineage F, but neither difference was significant.
Table 5. t-test of differences in the count of each clonal lineage between pairs of hosts.
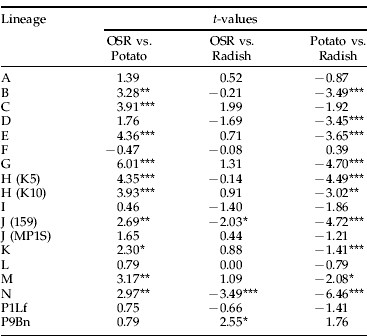
t-tests were used to determine if differences in lineage count between hosts were significant.
* =P<0.05; **=P<0.01; ***=P<0.001; df=63. Results from P floridana are not shown; but, for almost all comparisons, the colonies on this plant were significantly smaller.
The Pearson's correlation was calculated for the count of each clonal lineage on pairs of host plants. There was a significant positive correlation between OSR and radish (r=0.74, P<0.01). There was a positive correlation between count on potato and OSR, but it was not significant (r=0.55, P>0.02). Removing clonal lineage F reduced the r value of both results, but the correlation between OSR and radish remained significant.
Comparison between Scottish and Australian clonal lineages
Using a brassica host (cabbage, Brassica oleracea), Vorburger (Reference Vorburger2005) yielded measurements of reproductive rates that were very similar to those we found on OSR. Vorburger (Reference Vorburger2005) measured separate reproductive parameters, but it is still possible to calculate the 15-day colony count for his clones from the data presented. There is an excellent correlation (r=0.92, P<0.001, 17 d.o.f.) between the 15-day colony count from Vorburger's (Reference Vorburger2005) experiments with his most significant life-table measurement, ‘daily fecundity 2′. The range of counts calculated for the Australian clone lineages in Vorburger (Reference Vorburger2005) is 34–50, with an average overall lineage of 43. Our count results on the brassica oilseed rape are within two units, as they range from 32 to 52, after removing clone F, which was clearly a very poor performer and an outlier. The average colony count over all lineages of our results was 41 (again excluding clone F). The results of both the current and Vorburger's (Reference Vorburger2005) study using brassica plants as hosts are clearly in very close agreement. Vorburger (Reference Vorburger2005) concluded that his experimental conditions, including the use of cabbage, resulted in good growth and low mortality. This agrees with our conclusion that brassica plants appear to be optimal hosts for M. persicae.
Comparison between Scottish and German clonal lineages
The results we obtained for potato and OSR were in agreement with Weber (Reference Weber1985), who found that OSR produced the best aphid reproductive performance averaged over all lineages, followed by potato and then sugar beet. It is not possible to make direct comparison with the absolute values of Weber (Reference Weber1985), but indirect comparison is possible. In our studies, the overall count of all clonal lineages on potato was 77% of the overall count for OSR. For Weber (Reference Weber1985), the equivalent measurement is the average fecundity and this was 1.679 on OSR and 1.421 on potato, and the latter is 85% of the former. Thus, the relative performance over all the M. persicae lineages on potato and OSR are similar in the two studies. It was also possible to compare the performance of the best and worst performing lineages on potato and OSR in both studies. For OSR, there was very close agreement, with the worst performing lineage being at 35% of the best in our work and at 34% in Weber (Reference Weber1985). The same measurement was not as close on potato (57% in this study and 28% in Weber (Reference Weber1985)). Thus, despite these studies being conducted at different times and different regions, they do support each other in terms of the range and relative performance of M. persicae clonal lineages on the same plants. Weber (Reference Weber1985) concluded that he was working with a heterogeneous population. It does seem unlikely that he was examining a restricted set of clones, as sexual morphs and genetic variation have been recorded in M. persicae populations close to the German border (Guillemaud et al., Reference Guillemaud, Mieuzet and Simon2003). His work was also being carried out before the major expansion of MACE clones (Foster et al., Reference Foster, Denholm and Devonshire2000; Kasprowicz et al., Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a).
Does lineage reproduction predict field numbers?
Vorburger (Reference Vorburger2005) suggested a correlation between reproductive fitness and ecological success of Australian M. persicae clones. In our study, a large count for a clone lineage did not necessarily correspond to a clone with greater long-term success in the field. When the overall frequency of a clone was plotted against the count of its lineage on the two field hosts, there was no significant correlation (OSR, r=0.044, P>0.05; potato r=0.08, P>0.05). For example, the lineages of clone J, one of the long-lived, dominant, insecticide sensitive clones (Fenton et al., Reference Fenton, Malloch, Woodford, Foster, Anstead, Denholm, King and Pickup2005) had a much lower count on OSR and radish than a lineage of clone G, which is also sensitive but one of the rarest clones in the field (Kasprowicz, Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a,Reference Kasprowicz, Malloch, Pickup and Fentonb). Despite this, a lineage of clone H had the largest count on OSR, potato and radish; and between 4 and 14% of field-collected M. persicae belonged to this clone, depending on the season (Kasprowicz et al., Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a). Clone H appears abundant in the field when the conditions are suitable, and its lineages also appeared to reproduce well. A lineage of clone F had the lowest count, and this clone has not been found in the field since 1995 when it consisted of 4.5% of the samples; so, in this case, the low reproductive rate on a number of hosts does appear to be linked to a lack of field success (Kasprowicz, Reference Kasprowicz, Malloch, Pickup and Fenton2008b). The long-term survival of both clones H and F is likely to be negatively influenced by their insecticide resistance characteristics, but in different ways. In the case of H, this is indirect, for example through increased vulnerability to predation; whereas, clone F appears to have incurred a direct reproductive cost. A balance between selection by insecticides for increased abundance of resistant clones in the field followed by selection against these clones is a factor in clone success (Foster et al., Reference Foster, Denholm and Devonshire2000).
Clonal lineage performance at 14°C
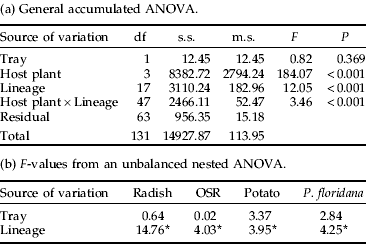
The counts of all 18 clonal lineages were measured on OSR at 14°C. Counts were significantly reduced when compared with those on OSR at 18°C, with the reduction averaged over all clonal lineages being around 80% (table 6). There was a significant interaction between temperature and clone (table 6). The largest count measurements at 14°C were observed for clone H lineages (K5 and K10), followed by I, N and then C. This resulted in their counts being reduced the least (fig. 3). The smallest count was produced by clonal lineages F, J (MP1S) and P1Lf with average colonies of 2–3 individuals. In the latter cases, many of the founding nymphs had not reached reproductive maturity, resulting in the greatest reduction in colony counts.

Fig. 3. The percentage of count when aphids are grown at 14°C compared to 18°C. The dotted line represents the average decrease in count across all clonal lineages (20%).
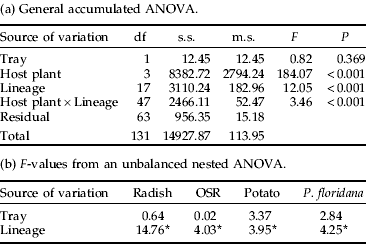
Table 6. A general accumulated ANOVA for count of M. persicae clonal lineages on OSR after 15 days at either 14°C or 18°C.
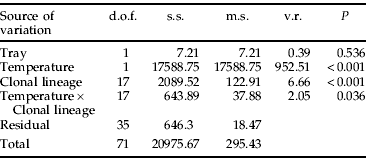
The count of all 18 clonal lineages was measured on OSR at 14°C and the values compared to those on OSR at 18°C using a general accumulated ANOVA. There was a significant interaction between temperature and clonal lineage. For abbreviations see table 2.
Pairwise comparisons found that the two pairs of lineages from clone H, K5 and K10 (t=0.32, df=1, P=0.753) and clone J, 159 and MP1S (t=1.70, df=1, P=0.098) produced similar counts at 14°C.
There was a positive correlation between the performance of a clonal lineage on OSR at 14°C and 18°C (r=0.64, P<0.01). This suggests that temperature is directly influencing aphid reproduction, as would be expected from insect physiology. However, not all clonal lineages were affected by the reduced temperature in the same way. A lineage of clone I, one of the dominant Scottish clones, had the lowest percentage decrease in count. This clone is more abundant in the northern regions, consistent with increased reproduction at lower temperatures and an advantage in the north (Kasprowicz et al., Reference Kasprowicz, Malloch, Pickup and Fenton2008b). This would also account for its disproportionate representation in suction traps, where it appears to have an extended season, i.e. it is present in the suction trap in colder months (Kasprowicz et al., Reference Kasprowicz, Malloch, Pickup and Fenton2008b). In contrast, a lineage of clone L, which is exclusively found in northern areas (Kasprowicz et al., Reference Kasprowicz, Malloch, Pickup and Fenton2008b) did not have a high count at the lower temperature. The other clonal lineages that reproduced well at lower temperatures belonged to clone C, which is a long-lived and widespread clone, and clone H, which had good reproductive rates in almost every experiment. Lineages of the other long-term parthenogenetic insecticide-sensitive clone J appeared to have a similar count at the lower temperature to most other clones.
General purpose genotypes and host associated genotypes
According to the general purpose genotype (GPG) hypothesis, environmental variation over time drives specialized genotypes of asexual organisms extinct; and, as a result, generalized genotypes are selected (Lynch, Reference Lynch1984). For aphid clonal lineages, clear factors in this extinction process will be the seasonal availability of herbaceous plant hosts and an ability to move to and from alternative hosts (Williams et al., Reference Williams, Dewar, Dixon and Thornhill2000). For M. persicae, this selection is believed to mean that the older a clonal lineage is, the more likely it is to be a GPG (Vorburger et al., Reference Vorburger, Sunnucks and Ward2003b). There was no evidence of GPGs in Australian M. persicae when asexual clonal lineages were grown on radish, spinach, tomato and lettuce (Vorburger et al., Reference Vorburger, Sunnucks and Ward2003b).
Clonal selection is likely to be strongest on hosts where reproduction is lowest and increased reproduction has the greatest advantage. In our study, stronger selection would occur on P. floridana and potato than on OSR and radish, although P. floridana does not occur locally. There was a good correlation between lineage growth on the two optimal hosts, but no correlation between lineage growth on the poor hosts (potato and P. floridana) or growth on the poor hosts and good hosts. The correlation suggests that, on good hosts, the lineages are less influenced by host plant biochemistry than by their own innate reproductive potential; and they are at, or close to, their maximum natural reproductive potential. On poorer hosts where correlations are weak or not found, more specific adaptations, probably linked to the presence or absence of biochemicals such as glycoalkaloids (GAs: Fragoyiannis et al., Reference Fragoyiannis, McKinlay and D'Mello1998) are involved. This will lead to a more complex interaction between clone genotype, reproductive performance and host-plant and insect biochemistry.
Although there was a host influence on the count of Scottish M. persicae clones, a growing colony could always be established on the plants we used, which indicates OSR, radish and potato are all potential field hosts. P. floridana produced the smallest colonies but it is not a field host.
A lineage of clone F produced essentially equivalent counts on all four hosts; and, despite these counts being small, this clone appears to be a GPG for these hosts. Clone L also appears to have the potential to be described as a GPG in Scotland, as its lineage produced similar colony counts on radish, OSR and potato, and this was also found to be a generalist in field studies (Kasprowicz et al., Reference Kasprowicz, Malloch, Pickup and Fenton2008b). It is interesting that clone L is a generalist, as it is totally restricted to the Elgin area (Kasprowicz et al., Reference Kasprowicz, Malloch, Pickup and Fenton2008b). Thus, the geographical restriction is not due to an inability to colonise a range of hosts.
Kasprowicz et al. (Reference Kasprowicz, Malloch, Pickup and Fenton2008b) showed that clone E was significantly over-represented on brassicas in Scottish field collections. Clone E may be relatively less abundant on potato, as it is unable to detoxify the plant's glycoalkaloids (GAs), which can suppress aphid reproduction (Fragoyiannis et al., Reference Fragoyiannis, McKinlay and D'Mello1998), but its performance on potato in the laboratory suggested it was not poorly adapted to this plant. This lineage of E had the third largest count on OSR. A lineage of C, another common clone, had good reproduction on OSR, and this clone shows a trend of increased abundance on brassicas in the field (Kasprowicz et al., Reference Kasprowicz, Malloch, Pickup and Fenton2008b). Clone E and possibly C appear to be closer to being specialized to brassicas than the other clones. Lineages of clones H and J also have good reproduction rates on brassicas in our current experiments but show no overrepresentation on this crop in the field (Kasprowicz et al., Reference Kasprowicz, Malloch, Pickup and Fenton2008b); and, therefore, growth and reproduction on brassica alone do not appear to account for the field distribution of clone E.
Clone J, lineage 159, produced larger colonies than lineage MP1S on three out of four of the equivalent host plants, and on radish this was significant. Lineage 159 was isolated in 2001, whereas lineage MP1S was isolated in 1977 and has been maintained in the laboratory ever since. The better growth of lineage 159 may reflect the continued selection for adaptability under field conditions.
The results between, and even within, clones suggests that the M. persicae population does contain clonal lineages with different host performance abilities, which reflect different levels of specialisation. However, all clonal lineages we tested have an ability to grow on all of the host plants to a greater or lesser degree. The most noted case of M. persicae host specialisation are the tobacco feeding forms, which are morphologically and genetically distinct from other populations, but this distinction has to be maintained by selection on tobacco (see Margaritopoulos et al., Reference Margaritopoulos, Kasprowicz, Malloch and Fenton2009).
A mixture of generalist and specialist genotypes has been seen in Chilean field populations of S. avenae where some of the genotypes were found across all hosts in equal frequencies and others were over-represented on different hosts (Figueroa et al., Reference Figueroa, Simon, Le Gallic, Prunier-Leterme, Briones, Dedryver and Niemeyer2004). But, as with the Scottish populations of M. persicae, no single dominant clone was collected from only one host plant species. In Mexican populations of B. brassicae, different clones were associated with the brassica hosts. Brassica campestris L. compared with B. oleraceae L. (Ruiz-Montoya et al., Reference Ruiz-Montoya, Núñez-Farfan and Vargas2003), but the differences were not extreme.
In the pea aphid, A. pisum, host specialisation is more extreme than in M. persicae (Leonardo & Muiru, Reference Leonardo and Muiru2003) and this has been attributed to nuclear genes (Hawthorn & Via, 2001). There is also evidence that secondary symbionts may play a role in plant colonisation (Leonardo & Muiru Reference Leonardo and Muiru2003). Secondary symbionts cannot be offered as an explanation for host plant performance of M. persicae, as their frequency in this aphid species is extremely low and they have not been implicated in this role (Chen & Purcell, Reference Chen and Purcell1997; Russell et al., Reference Russell, Latorre, Sabater-Muñoz, Moya and Moran2003; von Burg et al., Reference von Burg, Ferrari, Muller and Vorburger2008).
Insecticide resistance
The most obvious factor that could affect a clone's fitness is the insecticide resistance mechanisms it carries. Eggers-Schumacher (Reference Eggers-Schumacher1983) found that, under favourable conditions, clones with R1 or R2 esterase had a higher reproductive performance than sensitive clones on excised potato leaves. To balance this, susceptible clones appeared to be able to colonise less suitable host plants (Eggers-Schumacher, Reference Eggers-Schumacher1983). Other studies have shown that a high level of esterase is detrimental to a M. persicae clone's overwintering ability (Foster et al., Reference Foster, Harrington, Devonshire, Denholm, Devine and Kenward1996, Reference Foster, Harrington, Devonshire, Denholm, Clark and Mugglestone1997). Carrying kdr can alter an aphid's ability to react to alarm pheromones and, as a result, avoid parasitoid attack (Foster et al., Reference Foster, Woodcock, Williamson, Devonshire, Denholm and Thompson1999, Reference Foster, Kift, Baverstock, Sime, Reynolds, Jones, Thompson and Tatchell2003, Reference Foster, Denholm, Thompson, Poppy and Powell2005). The association between insecticide resistance and reproductive rates has been investigated using r m (intrinsic rate of increase) in different clones. The presence of elevated esterase levels and MACE in a clone appeared to reduce its r m (Foster et al., Reference Foster, Kift, Baverstock, Sime, Reynolds, Jones, Thompson and Tatchell2003). As a result, it was hypothesized that insecticide-sensitive Scottish clones would have a larger count than resistant clones. However, in the current study, there was no clear pattern between the count of lineages of sensitive and resistant clones. Clone H had the best reproductive performance averaged over potato, OSR and radish, yet this clone has MACE and kdr insecticide resistance. The insecticide-sensitive clone G was a very good performer as was the lineage of clone E, which carries R3 esterase. R3 esterase levels have been implicated in reduced r m (Foster et al., Reference Foster, Denholm, Thompson, Poppy and Powell2005), but this does not appear to be the case for E. Clone F, which is also esterase R3, but is also the only clone homozygous for kdr in this study, did appear to suffer a penalty for resistance as its lineage had the lowest reproduction on all host plants. Homozygous kdr genotypes like clone F are very rare in the field, even in mating populations where the R allele is reasonably frequent (Anstead et al., Reference Anstead, Mallet and Denholm2007), suggesting there is a direct and strong fitness cost associated with the resistant homozygote. We can now suggest that this could be caused by low reproduction, although we only have one example of this genotype.
Interestingly, lineages of the three most recent MACE clones to appear in Scotland, H, M and N (Kasprowicz et al., Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a), had higher counts than lineages of clones A and B, the two MACE-carrying clones that colonized in 2001 but subsequently disappeared. The MACE clones have exhibited a turnover (Kasprowicz et al., Reference Kasprowicz, Malloch, Foster, Pickup, Zhan and Fenton2008a), and this could now be accounted for by the more recent clones having evolved increased reproductive potential as an immediate compensation for other fitness costs.
Conclusions
The overall Scottish M. persicae population includes considerable numbers of insecticide-sensitive or non-MACE-resistant clones. These clones appear to have an advantage that we have now demonstrated is not overtly due to an increase in reproductive potential. Other factors measured in this study that could account for their relative success include: better growth rates at lower temperatures (clones I and C); better growth rates on brassicas (clones E and C); and insecticide resistance combined into a suitable genetic background (E and C). Another group of clones of more recent origins carry insecticide resistance including MACE. Factors known to affect survival of insecticide-resistant genotypes include reduced survival over winter (Foster et al., Reference Foster, Harrington, Devonshire, Denholm, Devine and Kenward1996) and altered pheromone responses leading to increased susceptibility to parasitism (Foster et al., Reference Foster, Denholm, Thompson, Poppy and Powell2005). We have now demonstrated that insecticide-resistant clone's growth rates vary but can be equal or better than fully susceptible clones. Our results suggest that initial fitness costs associated with insecticide resistance could be overcome by increased reproductive rates, as the two are not directly linked. Over time, genetic recombination and selection will generate insecticide resistant genotypes that are optimally adapted to the complex of positive and negative factors they encounter. Some of these will emerge as insecticide-resistant superclones and pose an ever increasing problem for agriculture.
Acknowledgements
This work was supported by a policy flexible fund project grant from the Scottish Government Rural and Environmental Affairs Directorate (RERAD). The authors wish to thank colleagues at SCRI and Rothamsted Research, particularly Alison Lees, Dave Cooke, Steve Foster and Ian Denholm for helpful discussions, as well as the many growers and agronomists who helped provide aphid material. Finally, thanks to Philip Smith for proofreading, Christine Hackett and Mark Phillips of Biomathematics and Statistics Scotland for statistical analysis and the Scottish Society for Crop Research for their support.