Introduction
Thrips (order Thysanoptera) are small insects (average length: 1–2 mm) with a global distribution. More than 5500 thrips species have been identified (Mound, Reference Mound and Lewis1997; Morse & Hoddle, Reference Morse and Hoddle2006). Some are considered pests that result in losses to the agriculture, horticulture, and forestry industries by directly damaging plants or through the transmission of tospoviruses (Moritz et al., Reference Moritz, Delker, Paulsen, Mound and Burgermeister2000). The most harmful thrips species are Frankliniella occidentalis (Pergande) and Thrips palmi Karny (Seal et al., Reference Seal, Kumar, Kakkar and Mello2013), though others, including Thrips tabaci, are also economically important.
Melon thrips [i.e., T. palmi Karny (Thysanoptera, Thripidae)] are polyphagous insects that feed on Cucurbitaceae and Solanaceae plant species (Walker, Reference Walker1994). Lewis (Reference Lewis1997) compiled a list of more than 200 plants on which T. palmi has been detected, including eggplant, cucumber, and watermelon. Other susceptible crops include pepper, sesame, sunflower, soybean, cowpea, tobacco, and squash. T. palmi was discovered in Indonesia, and has since spread to other regions with tropical and subtropical climates (Smith et al., Reference Smith1997). An outbreak of T. palmi destroyed almost 80% of the watermelon plantations in two regions of the Philippines in 1977. In 1978, this pest became a major threat to vegetable production in Japan, and during the subsequent decade, it spread to about 20,000 ha (Murai, Reference Murai2002). T. palmi is also present in South America, where it was reported for the first time in Venezuela infesting bean, potato, eggplant, melon, and other crops (Cermeli & Montagne Reference Cermeli and Montagne1993). In Australia, T. palmi was first observed on watermelon and cucumber (Houston et al., Reference Houston, Mound and Palmer1991), while in Europe, it was detected in glasshouses in the Netherlands in 1988 and 1992 (Mound & Gillespie, Reference Mound and Gillespie1997). In both cases, the pest was eradicated by destroying all infected plants (Cannon et al., Reference Cannon, Matthews and Collins2007). T. palmi adults and larvae feed on leaves and stems, which are attacked at or near the growing tip. They also occur on fruit surfaces and on petals and developing ovaries. The estimated economic losses in the UK resulting from infestations of greenhouse-grown plants by T. palmi are considerable. Additionally, a benefit-to-cost ratio for one eradication campaign was calculated as 110:1 (MacLeod et al., Reference MacLeod, Head and Gaunt2004). Heavy T. palmi infestations of plants in Florida, USA were associated with decreases in crop marketability. Moreover, commonly used insecticides alone or in combination are unable to satisfactorily control T. palmi during the early stages of infestation (Seal et al., Reference Seal, Kumar, Kakkar and Mello2013). In Taiwan, the market value of T. palmi-damaged fruits was only approximately 25% of that of undamaged fruits (Yadav & Chang, Reference Yadav and Chang2012).
F. occidentalis (i.e., western flower thrips) is another harmful thrips species. It has a very wide host range that spans horticultural and ornamental crops as well as trees, with hosts in 65 families (CABI, 2016). Alfalfa, chrysanthemum, corn, cotton, cucumber, eggplant, gerbera, gladiolus, grapes, impatiens, melons, peanut, pepper, plums, strawberry, tomato, and watermelon are among the plant species susceptible to F. occidentalis infestations (Chau & Heinz, Reference Chau and Heinz2006; CABI, 2016). F. occidentalis is one of the most important insect pests affecting glasshouse crops worldwide (Cloyd, Reference Cloyd2009), and it is also a major pest of some outdoor crops in warm climates. It is considered one of the most invasive species in Europe and can cause yield losses of up to 70% in greenhouse-grown cucumber (Marullo, Reference Marullo, Marullo and Mound2002). The effects are more serious when thrips populations function as virus vectors. F. occidentalis is endemic to North America, but it was also detected in the Netherlands in 1983, from where it spread across Europe (Kirk & Terry, Reference Kirk and Terry2003). It was subsequently detected in eastern Africa, New Zealand, and Australia. F. occidentalis occurs in many different habitats (e.g., lowland to alpine). However, because it does not undergo developmental or reproductive diapause (Ishida et al., Reference Ishida, Murai, Sonoda, Yoshida, Izumi and Tsumuki2003), F. occidentalis may not survive cold winters (McDonald et al., Reference McDonald, Bale and Walters1997). According to CABI (2016), F. occidentalis is currently distributed in several countries in Asia, Africa, North America, South America, the Caribbean, Europe, the Mediterranean, and Oceania. Importantly, the geographic distribution of F. occidentalis may overlap with that of T. palmi. Welter et al. (Reference Welter, Rosenheim, Johnson, Mau and Gusukuma-Minuto1990) studied mixed infestations of these two pests under experimental conditions, and observed significant decreases in total cucumber yield as well as mean and total fruit sizes.
F. occidentalis and T. palmi are vectors of serious plant viruses, including some of quarantine importance. T. palmi transmits several plant viruses, including watermelon silver mottle virus (Iwaki et al., Reference Iwaki, Honda, Hanada, Tochihara, Yonaha, Hokama and Yokoyama1984; Riley et al., Reference Riley, Joseph, Srinivasan and Diffie2011), tomato spotted wilt virus (Fujisawa, Reference Fujisawa1988), calla lily chlorotic spot virus (Chen et al., Reference Chen, Chen, Lin, Yeh and Hsu2005), groundnut bud necrosis virus (Reddy et al., Reference Reddy, Ratna, Sudarshana, Poul and Kumar1992), and melon yellow spot virus (Kato, Reference Kato2000). Watermelon silver mottle virus is on the European and Mediterranean Plant Protection Organization (EPPO) A1 list of pathogens to be regulated as quarantine pests (EPPO, 2015a), and causes severe losses to watermelon production in Taiwan (Yeh et al., Reference Yeh, Lin, Cheng, Jih, Chen and Chen1992), Japan (Okuda et al., Reference Okuda, Takeuchi, Taba, Kato and Hanada2002), and China (Rao et al., Reference Rao, Liu, Wu and Li2011). F. occidentalis is a vector of tomato spotted wilt virus, tomato chlorotic spot virus, and groundnut ringspot virus or impatiens necrotic spot virus (Wang et al., Reference Wang, Lin, Chiu and Shih2010). Worldwide crop damage caused by tospoviruses transmitted by F. occidentalis is likely greater than US$1 billion per year (Goldbach & Peters, Reference Goldbach and Peters1994). Tomato spotted wilt virus is a particularly important tospovirus that is transmitted by both T. palmi and F. occidentalis. It was first reported in tomato, but has been detected in approximately 900 host plants from 80 families. It has a worldwide distribution because its thrips vectors are widespread (Boonham et al., Reference Boonham, Smith, Walsh, Tame, Morris, Spence, Bennison and Barker2002). In the European Union, T. palmi is on the EPPO A1 list, while F. occidentalis is on the EPPO A2 list of pests that should be regulated as quarantine organisms (EPPO, 2015b).
Accurately identifying thrips species is crucial for managing these insect pests. It is also important to distinguish pests from non-pest species, and differentiate quarantine from non-quarantine insects. Identifying different thrips species based on morphological features requires entomological expertise, especially when distinguishing among larval species. However, molecular biology-based tools may enable the development of fast, easy, and sensitive diagnostic protocols for many pest species. There are methods for the molecular identification of T. palmi based on real-time polymerase chain reaction (PCR) detection (Kox et al., Reference Kox, Van Den Beld, Zijlstra and Vierbergen2005), sequence-characterized amplified region marker-based real-time PCR detection using a TaqMan probe (Walsh et al., Reference Walsh, Boonham, Barker and Collins2005), PCR–restriction fragment length polymorphism (RFLP) detection (Brunner et al., Reference Brunner, Fleming and Frey2002; Toda & Komazaki, Reference Toda and Komazaki2002), and loop-mediated isothermal amplification assays (Przybylska et al., Reference Przybylska, Fiedler, Kucharczyk and Obrępalska-Stęplowska2015). There are also protocols for identifying F. occidentalis using a real-time PCR assay with a TaqMan probe (Huang et al., Reference Huang, Lee, Yeh, Shen, Mei and Chang2010), PCR–RFLP (Brunner et al., Reference Brunner, Fleming and Frey2002; Toda & Komazaki, Reference Toda and Komazaki2002; Mainali et al., Reference Mainali, Shrestha, Lim and Kim2008; Przybylska et al., Reference Przybylska, Fiedler and Obrępalska-Stęplowska2016), and a cytochrome oxidase I mitochondrial DNA marker (Zhang et al., Reference Zhang, Meng, Min, Qiao and Wan2012) with a duplex PCR (Zhang et al., Reference Zhang, Wu, Zhou, Meng and Wan2014). Methods are also available describing a multiplex PCR assay for F. occidentalis, Frankliniella intonsa, T. tabaci, and T. palmi (Nakahara & Minoura, Reference Nakahara and Minoura2015) as well as a multiplex PCR assay for F. occidentalis, F. intonsa, T. tabaci, and Thrips hawaiiensis (Yeh et al., Reference Yeh, Tseng, Chang, Wu and Tsai2014). However, a protocol for detecting and differentiating between T. palmi and F. occidentalis in a single real-time PCR has not been developed.
The aim of this study was to develop a fast and sensitive method for simultaneously identifying and differentiating between F. occidentalis and T. palmi in one tube. We developed a duplex standard and real-time PCR protocol based on a melting curve analysis of the amplified 5.8S–internal transcribed spacer 2 (ITS2) ribosomal DNA (rDNA) fragment.
Materials and methods
Thrips samples
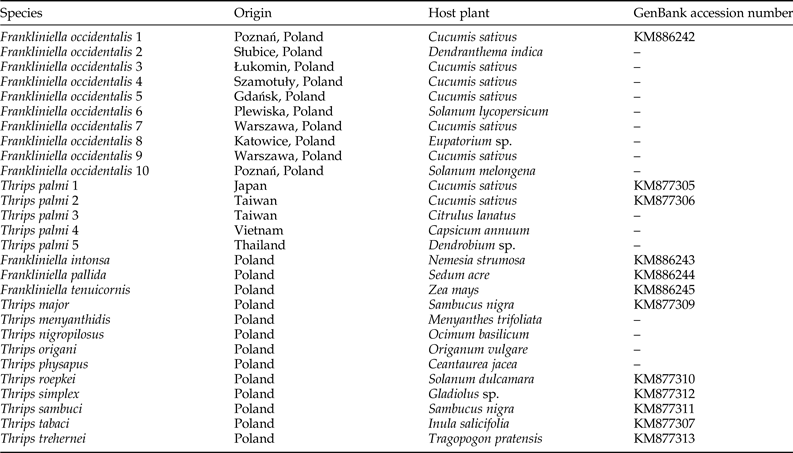
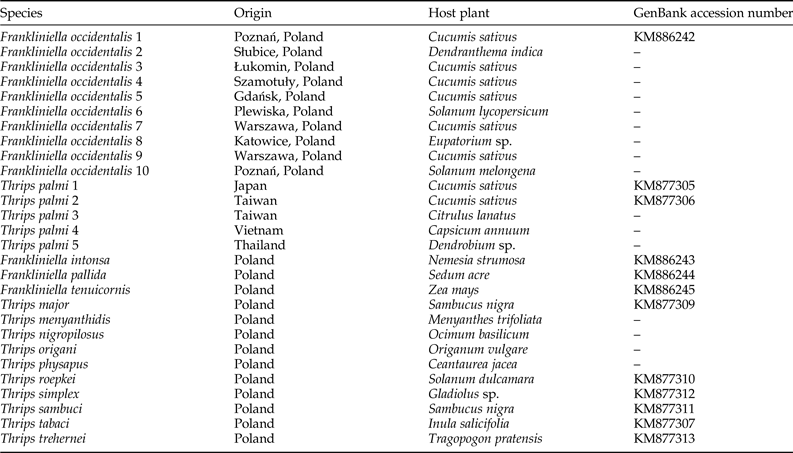
We examined ten F. occidentalis populations and six T. palmi populations. Additionally, single populations of T. major, T. menyanthidis, T. nigropilosus, T. origani, T. physapus, T. roepkei, T. simplex, T. sambuci, T. tabaci, T. trehernei, F. intonsa, F. pallida, and F. tenuicornis were used as negative controls. The geographical locations and host plants from which thrips specimens were collected are indicated in table 1.
Table 1. Thrips populations used as positive and negative controls in the study.
Genomic DNA extraction
Total genomic DNA was extracted from one, or in the case of some populations (derived from our previous study; Przybylska et al., Reference Przybylska, Fiedler, Kucharczyk and Obrępalska-Stęplowska2015), a few specimens, using the NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany) in a final volume of 100 µl. Final DNA concentrations were 5–10 ng µl−1. Genomic DNA from all populations which were taken as negative controls, were the same as these used in Przybylska et al. (Reference Przybylska, Fiedler, Kucharczyk and Obrępalska-Stęplowska2015), and during that study, they were tested in PCR with universal primers and sequenced.
Design of species-specific primers
Sequences used to design primers were obtained from Przybylska et al. (Reference Przybylska, Fiedler, Kucharczyk and Obrępalska-Stęplowska2015) and from a National Center for Biotechnology Information database. Sequences were aligned with the BioEdit program (Hall, Reference Hall1999), and a 5.8S rDNA fragment conserved in both species was used to design a universal forward primer (ThrUNIFw: 5′-GTGAACTGCAGGACACATG-3′). For F. occidentalis and T. palmi, regions in the ITS2 rDNA fragment lacking intraspecific and high interspecific variability were used to design species-specific reverse primers (FoRw: 5′-CGTAAACGACAGAACAG-3′ and TpRw: 5′-GCAGAGACACATCGCAAC-3′).
PCR assay
The primers were tested in PCR amplifications of F. occidentalis and T. palmi samples, in which DNA templates from both species were included in an equimolar ratio in one sample. We also completed PCR amplifications for the other thrips species (table 1), which served as negative controls. Samples with no genomic DNA were used to assess whether the reagents were contaminated. The PCR amplifications involved the following three primers sets: ThrUNIFw/FoRw to detect only F. occidentalis species, ThrUNIFw/TpRw to detect only T. palmi species, and ThrUNIFw/FoRw/TpRw to simultaneously detect both thrips species.
The singleplex PCR amplifications were completed with samples containing 10 ng template DNA, 0.5 µM forward and reverse primers, 5 µl DreamTaq master mix (Thermo Scientific, Waltham, USA), and sterile distilled water up to 10 µl. Duplex PCR amplifications were conducted with samples containing 10 ng template DNA, 0.33 µM of each of three primers, 5 µl DreamTaq master mix (Thermo Scientific), and sterile distilled water up to 10 µl. The PCR program was as follows: 95°C for 3 min; 30 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s; 72°C for 5 min. All amplified products were electrophoretically separated on a 1.5% agarose gel and visualized with Midori Green stain (Nippon) under UV light.
Duplex real-time PCR assay
A duplex real-time PCR assay was conducted for the F. occidentalis and T. palmi populations, with the other thrips species used as negative controls (table 1). A sample with no genomic DNA was used to determine whether reagents were contaminated. The results were verified based on melting curve analyses. Samples consisted of 1 µl template DNA (10 ng), 0.33 µM primers (i.e., three primers), 5 µl iTaq master mix (Bio-Rad, California, USA), and sterile distilled water up to 10 µl. The real-time PCR was conducted using a LightCycler 96 system (Roche) and the following program: 95°C for 5 min; 40 cycles of 95°C for 10 s, 54°C for 10 s, and 72°C for 10 s. The melting phase was initiated at 65°C and was completed at 95°C, with an increase of 1°C per step. To test the sensitivity of the PCR, a series of tenfold dilutions (starting from 10 ng µl−1 genomic DNA) of F. occidentalis and T. palmi templates was examined by real-time PCR as described above.
Results
Detection of F. occidentalis and T. palmi using singleplex and duplex PCR assays
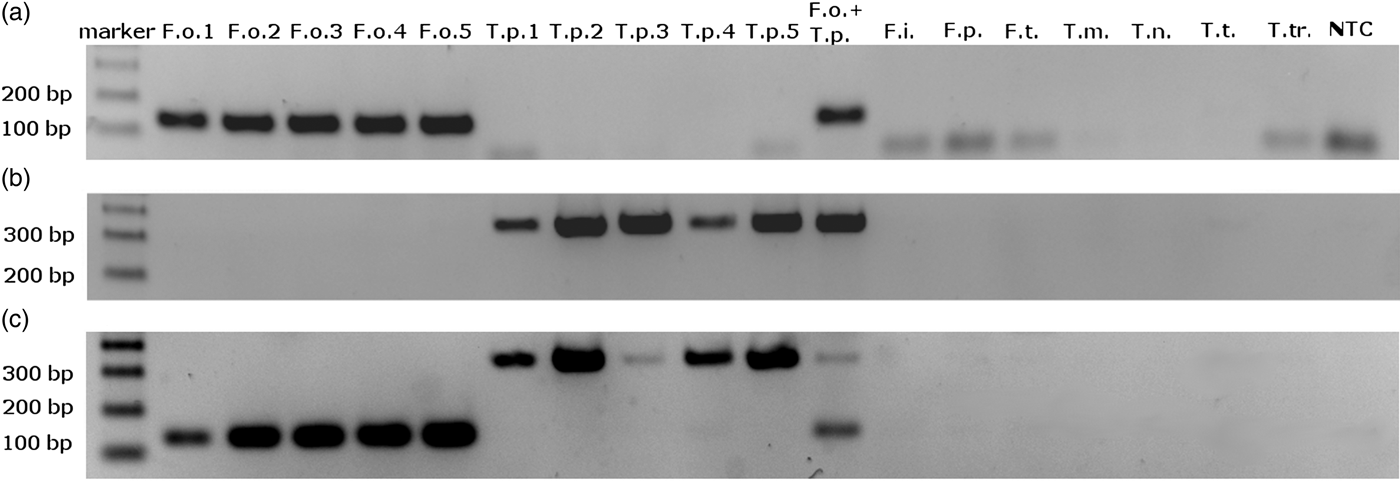
We obtained single amplification products for F. occidentalis and T. palmi samples using the ThrUNIFw/FoRw and ThrUNIFw/TpRw primer pairs, respectively. Amplifications with the ThrUNIFw/FoRw/TpRw primer set produced bands corresponding to the expected sizes of both thrips species. Amplicons were not detected for the other thrips species (table 1) and for the control sample lacking genomic DNA (fig. 1a–c). Product sizes were 128 bp for F. occidentalis and 333 bp for T. palmi.
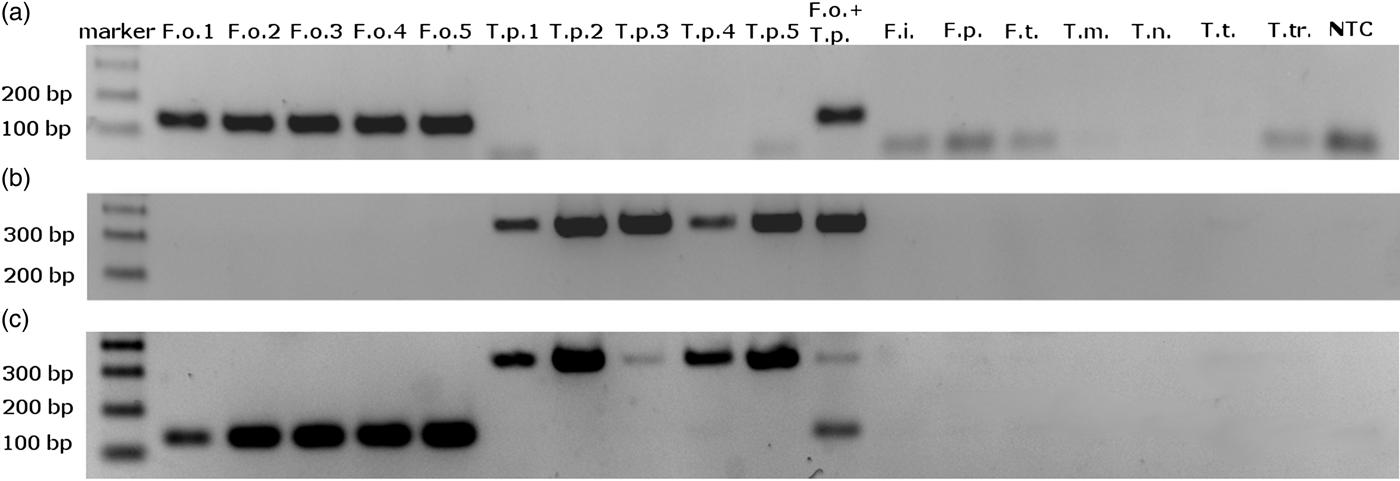
Fig. 1. Electrophoretic separation of PCR amplification products obtained with (a) ThrUNIFw/FoRw primers pair; (b) ThrUNIFw/TpRw primers pair; (c) ThrUNIFw/FoRw/TpRw primers set. DNA used to PCR were isolated from: F.o. – Frankliniella occidentalis, T.p. – Thrips palmi, F.i. – F. intonsa, F.p. – F. pallida, F.t. – F. tenuicornis, T.m. – T. major, T.n. – T. nigropilosus, T.t. – T. tabaci, T.tr. – T. trehernei; NTC – no template control.
Detection of F. occidentalis and T. palmi using a real-time PCR assay
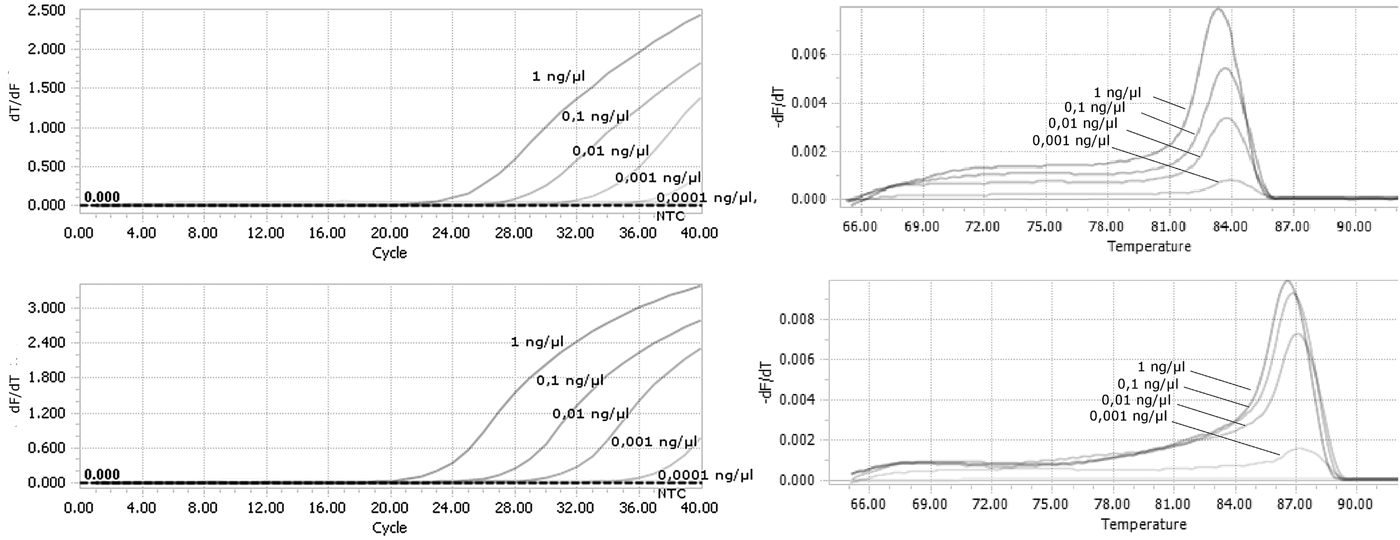
A duplex real-time PCR assay involving the ThrUNIFw/FoRw and/or ThrUNIFw/TpRw primer sets enabled us to distinguish between F. occidentalis and T. palmi species. Amplification and derivative melt curves were generated for F. occidentalis and T. palmi (fig. 2a, b), with a melting peak temperature range of 83.5–84.0°C for F. occidentalis and 85.5–87.0°C for T. palmi. No amplification products were produced for the other thrips species (table 1) or the control sample lacking genomic DNA.

Fig. 2. Results of real-time PCR amplification for all tested species. (a) Amplification curves; (b) Dissociation curves with melting peak temperature range between 83 and 83.5°C for F. occidentalis populations and between 85.5 and 87°C for T. palmi populations.
Tenfold serial dilutions of DNA isolated from F. occidentalis and T. palmi were used to evaluate the sensitivity of the detection assays. Our method was sufficiently sensitive to detect genomic DNA at a final concentration as low as 0.001 ng µl−1 for both species (fig. 3a, b). There were still observed derivative melt curves that matched the correct species in that concentration.
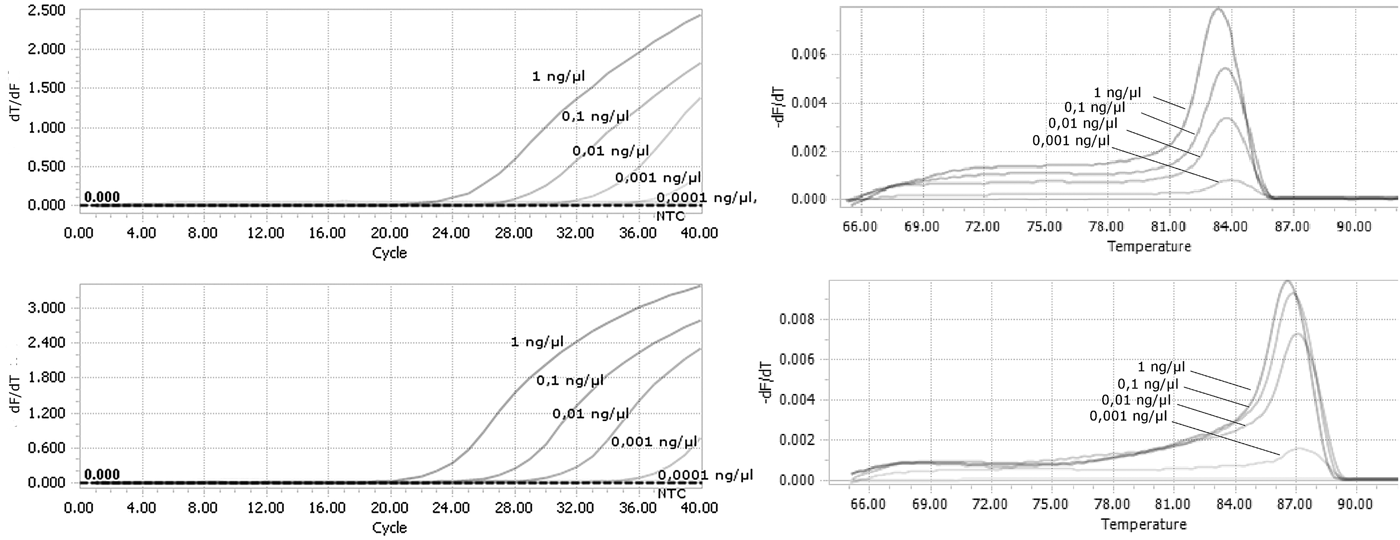
Fig. 3. Analysis of the sensitivity of real-time PCR detection. Genomic DNA from (a) F. occidentalis; (b) T. palmi were used as a template starting from the concentration of 1 ng µl−1 in final reaction mixture.
Discussion
The method developed in this study proved to be species-specific, fast, and sensitive. We used a wide array of thrips species closely related to T. palmi and F. occidentalis as negative controls. Przybylska et al. (Reference Przybylska, Fiedler, Kucharczyk and Obrępalska-Stęplowska2015) conducted a phylogenetic analysis based on rDNA fragments from many thrips species, and confirmed a close phylogenetic relationship between T. palmi and T. nigropilosus. Other closely related species are T. simplex, T. tabaci, and T. trehernei. Regarding F. occidentalis, all tested Frankliniella species clustered together in the phylogenetic tree. Thus, developing tests involving DNA samples isolated from several thrips species as negative controls is important because many of these species have common hosts and/or geographic distributions. However, most of the included thrips species do not affect crop production. No amplification product was observed for any of the non-target species during the singleplex PCR or real-time PCR assays. Moreover, PCR amplifications were completed with a mixture of F. occidentalis and T. palmi DNA or a mixture of their species-specific primer sets to assess reaction specificity. Our results indicated that the developed assay was species-specific, and able to distinguish between F. occidentalis and T. palmi in a single sample. Our method was sufficiently sensitive to detect genomic DNA at concentrations as low as 0.001 ng µl−1. Furthermore, the protocol was completed quickly because both species were simultaneously detected in a single real-time PCR sample and differentiated based on the melting temperature of the amplification products, which eliminates the need for a gel electrophoresis step. Therefore, our method may be useful for routine analyses of insect samples collected from fields or in greenhouses. Our assay may also enable the monitoring of imported plant material to prevent the spread of T. palmi (i.e., quarantine species) throughout Europe.
While designing primers, we examined rDNA sequences based on experimental data and National Center for Biotechnology Information database records to identify regions conserved in T. palmi and F. occidentalis populations from various geographical locations and plant hosts. We did not detect significant differences in the 5.8S–ITS2 fragment sequences among the F. occidentalis and T. palmi populations. We analyzed 5.8S-ITS2 fragment with the length of about 460 nucleotides for F. occidentalis with differences in length between populations not more than seven nucleotides and 580 nucleotides for T. palmi with no differences in length between all analyzed populations. There were some single nucleotide substitutions observed within amplified region of each species (2–15 different nts between analyzed F. occidentalis populations and 1–21 nts for T. palmi populations), but it should not have influence on primer hybridizations, PCR amplification, PCR product length, or the melting temperature of real-time PCR products. Thus, we assume that our method may be applied for detecting T. palmi and F. occidentalis populations regardless of their origin.
T. palmi and F. occidentalis are highly polyphagous pests with overlapping host ranges. The most important common plant hosts are cucumber, melon, and watermelon. These species can occur together in greenhouses or under field conditions in relatively warm countries where T. palmi occurs (CABI, 2016).
There are currently two PCR–RFLP assays for detecting and differentiating between T. palmi and F. occidentalis (Brunner et al., Reference Brunner, Fleming and Frey2002; Toda & Komazaki, Reference Toda and Komazaki2002). However, point mutations can generate false negative results in PCR–RFLP assays because restriction enzymes cut DNA at or near specific restriction sites and they are unable to recognize sites in which a nucleotide change has occurred. In contrast, PCR primers can hybridize properly even if there are single nucleotide mismatches between the primers and target sequence (Bru et al., Reference Bru, Martin-Laurent and Philippot2008; Stadhouders et al., Reference Stadhouders, Pas, Anber, Voermans, Mes and Schutten2010; Wieczorek & Obrępalska-Stęplowska, Reference Wieczorek and Obrępalska-Stęplowska2013). Moreover, a PCR–RFLP assay cannot be completed as quickly as duplex PCR or duplex real-time PCR assays because it involves two enzymatic reactions (i.e., PCR and RFLP) followed by an electrophoretic analysis. A multiplex PCR protocol is available for differentiating among F. occidentalis, F. intonsa, T. tabaci, and T. palmi (Nakahara & Minoura, Reference Nakahara and Minoura2015), but this method is not as fast as our real-time PCR protocol because it requires an additional step involving electrophoretic separation and visualization of amplification products. Furthermore, many previously published diagnostic protocols required the amplification of the cytochrome oxidase I mitochondrial DNA fragment. However, high intraspecific variability in this region has been reported for T. palmi and other thrips species (Rebijith et al., Reference Rebijith, Asokan, Kumar, Krishna and Ramamurthy2012). Therefore, we focused on a 5.8S–ITS2 rDNA fragment because Przybylska et al. (Reference Przybylska, Fiedler, Kucharczyk and Obrępalska-Stęplowska2015) revealed this region was relatively stable among examined populations. Moreover, part of the 5.8S rDNA fragment is conserved among many thrips species, enabling the use of a common forward primer for detecting F. occidentalis and T. palmi. In contrast, the ITS2 fragment is characterized by high interspecific sequence variability and thus, this region was ideal for designing species-specific reverse primers. In the majority of previously published protocols, the sensitivity was not tested or was assessed only for diluted larval or adult thrips samples. Thus, it is difficult to compare the sensitivity of our method with previously published procedures. Only Huang et al. (Reference Huang, Lee, Yeh, Shen, Mei and Chang2010) used tenfold dilutions of DNA samples starting from 10 ng µl−1. Their detection limit was 0.1 pg µl−1, suggesting their method is ten times more sensitive than our assay, but their protocol was designed to detect only F. occidentalis species.
The method described herein may be useful as a rapid test for the early and reliable detection of F. occidentalis and T. palmi, which may be relevant for increasing the quality and yield of agriculturally important crops.
Acknowledgements
The authors would like to thank Professor Tamotsu Murai from the Laboratory of Applied Entomology at Utsunomiya University, Japan; Dr Bert Vierbergen from Netherlands Food and Consumer Product Authority, Mr Chien-Hao Tseng from the National Taiwan University, Taipei, and Ms Yi-Ju Chen from the Applied Zoology Division, Taiwan Agricultural Research Institute, Taichung, for providing T. palmi material for the study. We also thank dr Halina Kucharczyk for providing F. tenuicornis, T. trehernei, T. nigropilosus, T. origami, T. physapus, T. roepkei, T. simplex, T. sambuci, and T. menyanthidis used as negative controls in this study. The study was supported by the Multiannual Program of the Polish Ministry of Agriculture and Rural Development (task 2.7).