Introduction
The order Thysanoptera comprises 7400 described species and nearly 8000 estimated species of small insects (0.5–3.0 mm), known as thrips, which present diverse life histories and a wide array of behaviors, ranging from solitary, subsocial to eusocial forms (Terry, Reference Terry and Lewis1997; Mound, Reference Mound2005). Two suborders, divided into nine subfamilies, are currently recognized: Terebrantia and Tubulifera, characterized by the presence and absence of an ovipositor, respectively (Mound & Morris, Reference Mound and Morris2007). These insects are considered important plagues whereas they can damage leaves, flowers and fruits while feeding, besides causing virus infections in plants (Mound & Marullo, Reference Mound and Marullo1996; Mound & Kibby, Reference Mound and Kibby1998).
Similarly to hymenopterans (bees, wasps and ants) and some coleopterans and homopterans (whitefly and aphids), thysanoptereans are haplodiploid insects and reproduce by parthenogenesis (Crespi, Reference Crespi, Wrensch and Ebbert1993; Kumm & Moritz, Reference Kumm and Moritz2008).
The first reports focusing on accessing the genetic variability of haplodiploid species were based on allozyme polymorphisms, but they proved to be unsuccessful because of their lack of variation (Snyder, Reference Snyder1974; Metcalf et al., Reference Metcalf, Marlin and Whitt1975; Pamilo et al., Reference Pamilo, Vepsalainen and Rosengren1975). Ever since, several hypotheses, mainly based on evolutionary genetics, were formulated to explain such apparent genetic homogeneity. The most acknowledged one refers to the effective population size that is expected to be three-quarters of that found in diploid species with equal sex ratio, thereby resulting in low heterozygosity levels in haplodiploid species (Hedrick & Parker, Reference Hedrick and Parker1997).
Over the last decade, molecular techniques have significantly contributed to the understanding of population genetics (Bertorelle et al., Reference Bertorelle, Bucchini, Pilastro and Matessi1999; Sunnucks, Reference Sunnucks2000). A high number of DNA markers have been developed and are now widely available for studies of genetic variation (Loxdale & Lushai, Reference Loxdale and Lushai1998; Bayar et al., Reference Bayar, Törjék, Kiss, Gyulai and Heszky2002). An advantage of certain DNA markers is their ability to examine the portions of an entire genome, thus allowing an increased knowledge about the genetic diversity of many loci. In population genetics, the RAPD (Random Amplification of Polymorphic DNA) technique is a useful tool (Jain et al., Reference Jain, Neekhra, Pandey and Jain2010). The RAPD technique has been applied in a variety of studies, such as genomic fingerprinting of individuals, lineages and populations, construction of genetic maps and localization of genes linked to economic traits (Welsh & McClelland, Reference Welsh and Mcclelland1990; Williams et al., Reference Williams, Kubelik, Livak, Rafalski and Tingey1990; Jain et al., Reference Jain, Neekhra, Pandey and Jain2010).
The RAPD technique has also been successfully applied in genetic studies of many insect species in order to differentiate geographically isolated populations or to verify genetic divergence influenced by selection under different environmental conditions or as a result of genetic drift (Fuchs et al., Reference Fuchs, Gross, Stein and Rottmann1998; Baumann et al., Reference Baumann, Schubert, Heitland, Auger-Rozenberg, Faivre-Rimpant and Müller-Starck2003; Borashi & Del Lama, Reference Boraschi and Del Lama2004; Callejas et al., Reference Callejas, Beitia, Gobbi, Velasco and Ochando2005). However, in haplodiploid insects, most molecular marker studies are focused on hymenopterans and little is known about other groups.
Reports about genetic variability and population structure in thrips are scarce. Therefore, in this work, RAPD markers were used to estimate both genetic variation and structuring in different populations of Gynaikothrips uzeli (Thysanoptera: Phlaeothripidae), as well as the role of haplodiploid sex system in genetic diversity.
Material and methods
Specimens of G. uzeli were collected in six localities, three populations from two cities in Paraná state; Ponta Grossa (PG1-25°5′47.74″S/50°12′17.18″W and PG2-25°6′4.19″S/50°12′6.48″W) and Curitiba (Ctb-25°26′55.61″S/49°13′57.98″W) and three from different municipalities in Bahia state: Itiruçu (Iti-13°32′1.42″S/40°9′1.52″W), Jequié (Jeq-13°51′42.98″S/40°5′17.67″W) and Jaguaquara (Jag-13°31′50.32″S/39°58′1.93″O) (fig. 1). The DNA extraction was performed according to Roberts (Reference Roberts1998). For each locality, 36 individuals were analyzed.

Fig. 1. (a) Geographic map of Brazil, showing the sampling sites of G. uzeli; (b) detail of Bahia state and collection cities; and (c) detail of Paraná state and collection cities.
A total of 30 RAPD primers were tested for the amplification of DNA fragments. Based on the amplification profile, reproducibility, number and resolution of bands, 14 primers were selected. The sequences and amplification pattern of each primer are presented in table 1.
Table 1. List of RAPD primers, showing their respective sequences, annealing temperature and number of bands.

The amplification reactions were carried out in a Gradient System Thermocycler (Biocycler). Each reaction comprised 15 μl of solution, containing: 1.5 μl of 10× buffer (75 mM Tris-HCl pH 9.0; 50 mM KCl); 4.0 mM MgCl2; 0.266 mM of each dNTP; 0.8 mM of primer; 1 U of Taq DNA polymerase (Invitrogen) and 30 ng of template DNA. For each primer, a negative PCR control was carried. The PCR involved one initial denaturation cycle at 94°C for 3 min, followed by 45 cycles at 94°C for 1 min, specific annealing temperature of each primer for 45 s, and extension at 72°C for 2 min plus a final extension step at 72°C for 6 min. The RAPD products were separated by electrophoresis in 2% agarose gel (p/v) for about 5 h at 50 V and stained with Gelred (Biotium) in 1:1000 dilution. The DNA fragments were visualized under UV light, and images were documented using MF-ChemiBis (DNR Bio-Imaging Systems Ltd, Jerusalem, Israel).
The RAPD fragments were visually scored according to their presence (1) or absence (0) among individuals. The software NTSYS-PC v 2.1 (Numerical Taxonomy and Multivariate Analysis System) (Rohlf, Reference Rohlf2000) was used for grouping analysis by UPGMA (unweighted pair group method with arithmetical average) (Sneath & Sokal, Reference Sneath and Sokal1973). The Jaccard's similarity coefficient was employed in this analysis. The bootstrap values were obtained with the WinBoot software (Yapi & Nelson, Reference Yapi and Nelson1996) using 1000 permutations.
Assuming Hardy-Weinberg equilibrium, the software POPGENE v. 1.32 (Yeh et al., Reference Yeh, Yang and Boyle1999) was used to estimate: gene frequency, Nei's gene diversity (He) (Nei, Reference Nei1973), Shannon index (Lewontin, Reference Lewontin1972), Nei's genetic distance and identity, and the percentage of polymorphic loci. The analysis of molecular variance (AMOVA) was performed to evaluate the genetic structure within and among populations according to Excoffier et al. (Reference Excoffier, Smouse and Quattro1992) in the software Arlequin v. 3.5 (Excoffier & Lischer, Reference Excoffier and Lischer2010). The structuring significance was tested using 1000 permutations.
Results
The amplification reactions using the 14 selected RAPD primers yielded 152 bands, ranging from 1500 to 100 base pairs. The mean number of bands per primers was equal to 11.2. The primers OPW-09 and OPE-18 produced the highest number of bands (16), while OPAV-13 amplified the lowest number (4) (table 1). Figure 2 shows the amplification pattern obtained with primer OPAV-3 in nine samples (each containing four individuals) from the six studied populations of G. uzeli.

Fig. 2. RAPD amplification pattern in 2% agarose gel of the six populations of G. uzeli using primer OPAV-3. Each lane represents the DNA amplified from a pool of four individuals. (M) 100 bp molecular weight marker.
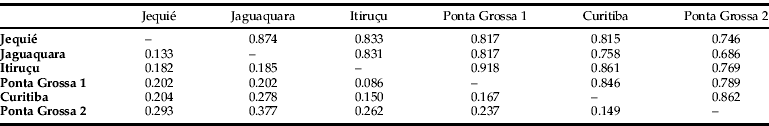
A total of 136 out of the 152 loci were polymorphic whereas 16 were monomorphic, resulting in a mean polymorphism of 89.47%. The values of Nei's gene diversity and Shannon index were 0.30 and 0.45, respectively. The values of pairwise Nei's genetic distance in the studied populations were reduced, being lower between the populations from Ponta Grossa 1 and Itiruçu (0.086) and higher (0.377) between Jaguaquara and Ponta Grossa 2 (table 2).
Table 2. Nei's genetic identity (above diagonal) and distance (below diagonal).
The UPGMA-based dendrogram invariably grouped all individuals within their respective collection sites (fig. 3). The coefficient of genetic similarity among populations ranged from 0.66 to 0.97. Considering a similarity coefficient of 67%, the grouping analysis revealed clusters: one comprising two populations from Bahia state (Jequié and Jaguaquara) and another, larger group comprising the populations from Paraná (Ponta Grossa 1 and 2, and Curitiba) and the population from Itiruçu (BA).

Fig. 3. UPGMA-based dendrogram based on genetic similarity using Jaccard's coefficient. Bootstrap values are given alongside the nodes (1000 replicates). Jeq, Jequié; Jag, Jaguaquara; Iti, Itiruçu; PG1, Ponta Grossa 1; Ctb, Curitiba; PG2, Ponta Grossa 2.
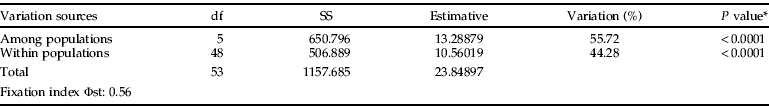
The AMOVA data performed in two hierarchical levels showed that populations are highly structured and that most of the genetic diversity was verified among populations (table 3).
Table 3. Analysis of molecular variance in six populations of Gynaikothrips uzeli, considering two hierarchical levels. df, degrees of freedom; SS, sum of squares; MS, mean square; estimative, estimative of variance components.
Discussion
Similarly to other haplodiploid insects, low levels of genetic diversity were observed in G. uzeli using RAPD markers. Such reduced variation might be related either to the haplodiploid sex determination or to ecological particularities of this species, like the low dispersal abilities and life cycle in leaf galls.
The haplodiploid sex determination leads to few polymorphic loci because of the strong selective pressure over recessive alleles in males, since all hemizygotic loci are exposed to selection and can be rapidly removed from the population (Hartl, Reference Hartl1971; Kerr, Reference Kerr1976; Hedrick & Parker, Reference Hedrick and Parker1997). The haplodiploidy also implies in decreased effective population size once, in species bearing this sex-determining mechanism, it equals three-quarters of the effective size values of diploid populations with equal sex ratio, thereby yielding comparatively low heterozygosity rates (Hedrick & Parker, Reference Hedrick and Parker1997). Moreover, a higher rate of allele fixation coupled with difficulties in keeping stable polymorphisms is reported in species bearing this system.
Nonetheless, some haplodiploid insects, such as the hymenopterans of the suborder Symphyta (Rosenmeier & Packer, Reference Rosenmeier and Packer1993; Boato & Battisti, Reference Boato and Battisti1996; Boraschi & Del Lama, Reference Boraschi and Del Lama2004), present heterozygosity levels similar to those reported in diploid species, suggesting that haplodiploidy per se should not be the only cause of restraining genetic variability within populations (Boraschi & Del Lama, Reference Boraschi and Del Lama2004). On the other hand, Crespi (Reference Crespi1991), based on isozyme analysis, found heterozygosity values in four species of thrips close to the rates reported in other haplodiploid insects, reinforcing the role of haplodiploid systems in determining low levels of genetic variation.
The high genetic similarity among studied populations may be associated with the short period of introduction of this species in Brazil (<100 years), which took place simultaneously to the importation of its host plant Ficus benjamina in 1920, and putative genetic drift effects that could potentially reduce the original gene pool in the introduced population. These factors, combined with the haplodiploidy, gall-associated behavior and dispersal constraints, could account for the low genetic variability reported in the studied species. Once the introduction of G. uzeli and the consequences of genetic drift could be recent events, the populations had little time to diverge significantly, thus resulting in high similarities even between geographically isolated populations such as Itiruçu and Ponta Grossa-1 (1662 km apart).
The flight ability of thrips is limited; thus, the individuals are usually dispersed by winds. Many species have reached new habitats via international trade of products such as fruits, vegetables and ornamental plants (Brunner et al., Reference Brunner, Fleming and Frey2002). Dispersal over short distances causes the populations to structure according to the isolation-by-distance model, as commonly reported for several organisms (Brunner & Frey, Reference Brunner and Frey2010). Therefore, it would be expected that populations of G. uzeli followed isolation-by-distance, in which closely located populations should be more similar than geographically distant ones.
However, the highest similarity levels were observed among samples of G. uzeli from similar habitats, as observed by the two distinct clusters in grouping analyses discriminating hot/dry from cold/humid habitats (table 4 and fig. 3). Brunner & Frey (Reference Brunner and Frey2010) reported a similar pattern in Frankliniella occidentalis using microsatellite and mitochondrial DNA markers. These results might reflect the adaptation of populations to similar environmental conditions. Nonetheless, this inference cannot be made in the present study, once the adaptive value of DNA regions amplified via RAPD is unknown (Waldschmidt et al., Reference Waldschmidt, Marco-Junior, Barros and Campos2000).
Table 4. Climate features of collection sites of populations of G. uzeli (adapted from IBGE, 2004).

The low genetic diversity in G. uzeli could also result from its life mode in leaf galls. Based on the hypothesis of niche variation (Van Valen, Reference Van Valen1965), which explains the polymorphism levels according to the species niche range, the heterozygosity usually increases in relation to environmental variation and decreases within more stable habitats. Inasmuch as galls, like colonies of hymenopterans, provide a relatively stable microenvironment, low levels of genetic diversity might be expected in this thrip species.
AMOVA among and within populations revealed that the overall genetic diversity is driven by inter-population differences (table 3). Although the studied populations are quite similar, AMOVA results showed that they are highly structured. Such structure level can be explained based on the haplodiploid system and the low dispersal abilities of G. uzeli.
One key force in the distribution of genetic diversity between and within populations is gene flow. The highest percentage of genetic variability within localities is putatively related to restricted (or absent) gene flow amongst individuals from distinct populations, determining a high inbreeding rate within localities and concentration of the low polymorphism generated by the haplodiploid system, thereby leading to population differentiation.
Further studies with other thysanopterans and haplodiploid insects, combining molecular, ecological and behavioral data, might eventually help elucidate the evolutionary role of the haplodiploid sex-determining system in the generation of genetic variability and its maintenance, in addition to providing a better understanding about the mechanisms involved in the dispersal and differentiation of species.
Acknowledgements
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq and by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-CAPES.









