Introduction
In contrast to invasive pests that originate from elsewhere and attack crops, native pest systems are typically viewed as stable, unchanging entities, and therefore often neglected for study. While methods for controlling native pests may draw research interest due to their economic impact, fundamental aspects of their biology and behaviour are often overlooked, and therefore poorly understood. Recent studies in California, USA, utilizing molecular methods, have cast doubt on species designations for some common native insect pests. For example, the California native western flower thrips Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae) was recently found to be comprised of two previously unrecognized, morphologically inseparable, cryptic species that occasionally co-occur on the same host plants (Rugman-Jones et al., Reference Rugman-Jones, Hoddle and Stouthamer2010). Similarly, the blue-green sharpshooter, Graphocephala atropunctata (Signoret) (Hemiptera: Cicadellidae), a native California pest of grapes, displays readily identifiable, population level variation in traits associated with communication, mating and genetic constitution, that may indicate incipient speciation (Ballman, Reference Ballman2009; Ballman et al., Reference Ballman, Rugman-Jones, Stouthamer and Hoddle2011). These findings are interesting from a biological perspective because they raise questions about the concept of ‘species’ and the speciation process itself, but they also have important economic ramifications since accurate species identification is critical in the control of pest arthropods (Rosen, Reference Rosen1986; Davies et al., Reference Davies, King and Smith2004; Armstrong & Ball, Reference Armstrong and Ball2005). Misidentification of a pest (or a natural enemy) may lead to the implementation of inappropriate control measures, resulting in considerable wasted time, money and effort (Rosen, Reference Rosen1986).
It is commonly accepted that speciation results from divergent natural and/or sexual selection acting on populations that occupy different environments (Turelli et al., Reference Turelli, Barton and Coyne2001; Coyne & Orr, Reference Coyne and Orr2004). In this sense, ‘environments’ may be ecologically different (e.g. different climate and host plants) and/or geographically different (i.e. allopatric). The key point is that, alleles which are advantageous (and therefore adaptive) in one environment may not be in another; and, as a result, selection (natural or sexual) drives the fixation of different alleles in the populations occupying those different environments. Given enough time, such divergent selection can incidentally lead to reproductive incompatibility between two populations (Muller, Reference Muller1942; Mayr, Reference Mayr1963). Common characteristics of the two native California pest examples given above, are: their apparent affinity for a wide range of host plants (polyphagy), potential for high reproductive output, which promotes pestiferousness and increases the potential for natural selection due to large population sizes, and broad ecological amplitude (the ability to occupy a wide variety of habitats with greatly differing climatic characteristics). Collectively, these attributes allow these native pests to exploit a broad niche range. Consequently, these conditions have provided opportunities for significant ecological and geographical differentiation between populations within each species, which, in the case of F. occidentalis, may have contributed to cryptic speciation and which may currently be contributing towards speciation in G. atropunctata.
Bean thrips, Caliothrips fasciatus (Pergande) (Thysanoptera: Thripidae), is native to North America, and is known principally from the western United States (US), although its distribution also extends eastward throughout the southern US to Florida and south into parts of western Mexico (Bailey, Reference Bailey1940; Hoddle et al., Reference Hoddle, Stosic and Mound2006, Reference Hoddle, Mound and Paris2008). Reports of C. fasciatus populations elsewhere are considered dubious (Hoddle et al., Reference Hoddle, Stosic and Mound2006; Mound et al., Reference Mound, Zhang and Bei2011). In California, C. fasciatus is widely distributed in urban, agricultural and natural areas. It can breed on various weed species including sowthistle (Sonchus oleraceus L.), cheeseweed (Malva parviflora L.) and fennel (Foeniculum vulgare Miller) and was once considered a serious pest of a variety of agricultural crops, including alfalfa, beans, cantaloupes, cotton, lettuce, pears, peas and walnuts (Bailey, Reference Bailey1933, Reference Bailey1937, Reference Bailey1938). The significance of this insect as an agricultural pest in California has waned substantially since Bailey's papers were published, perhaps due to the development of integrated pest management programs, use of more effective insecticides on crops that are most vulnerable to C. fasciatus outbreaks, development of resistant cultivars, and/or competitive exclusion by more aggressive species (e.g. F. occidentalis). However, C. fasciatus remains a major quarantine issue for the export of fresh navel oranges from California because this thrips may form aggregations in the navels of the oranges, thereby posing an incursion threat to importing countries (Hoddle et al., Reference Hoddle, Stosic and Mound2006; Australia Phyto Requirements 2010). Consequently, recent research efforts on C. fasciatus have focussed on detection (Harman et al., Reference Harman, Mao, Robinson and Morse2007a,Reference Harman, Mao and Morseb) and fumigation strategies (Leesch et al., Reference Leesch, Tebbets and Tebbets2004; Mitcham et al., Reference Mitcham, Bikoba, Pupin and Biasi2011; Morse et al., Reference Morse, Robinson and Urena2011) to reduce the frequency of contamination events. This focus on pragmatic solution-driven research has diverted attention from the basic biology of this pest. No studies have addressed biological attributes that could affect the invasion potential of C. fasciatus into countries that accidentally import California-grown navel oranges contaminated with the insect.
Given the propensity of C. fasciatus to occupy a wide variety of habitats and plant species (factors that could pre-dispose this pest to invasion), coupled with recent reports of cryptic speciation in species that share this affinity (e.g. F. occidentalis), a re-examination of the basic biology of the pest was thought pertinent. Primarily, we sought to qualify the specific status of C. fasciatus and then to identify characteristics that allow it to occupy such a wide range of habitats, which in turn may influence its ability to establish in countries importing navel oranges contaminated with this insect. One mechanism that could influence the ability of thrips to colonize new regions is their mode of reproduction. While the normal mode of reproduction in thrips is arrhenotoky, thelytoky is found in several species (Lewis, Reference Lewis1973) and, in some cases, is associated with infection by the endosymbiont bacterium Wolbachia (Pintureau et al., Reference Pintureau, Lassabliere, Khatchadourian and Daumal1999; Arakaki et al., Reference Arakaki, Miyoshi and Noda2001; Kumm & Moritz, Reference Kumm and Moritz2008). Thelytokous species are particularly prone to establish in new areas because only a single female is required to establish a population and no Allee effects hamper their spread. In addition, even virgin females of some arrhenotokous thrips species have been shown to produce low levels of female offspring (Kumm & Moritz, Reference Kumm and Moritz2010), which could improve establishment likelihood for such species.
Here, we report the findings of investigations into several aspects of the biology of Californian populations of C. fasciatus. First, we investigated if this species is genetically homogeneous or if it consisted of identifiable sub-groups, by determining genetic variation in the mitochondrial COI gene of individuals from populations across California. Second, we determined mating compatibility of three populations that differed in both the area in which they were collected and in their mitochondrial heritage (COI haplotype). Finally, we tested specimens for the presence of Wolbachia symbionts to determine if potentially thelytokous populations of this species exist. Endosymbiont mediated thelytoky could increase the invasion risk from C. fasciatus to countries that import California navel oranges accidentally contaminated with this thrips.
Materials & methods
Specimen collections and DNA extraction
Bean thrips were collected from a variety of host plants in areas throughout California (table 1). Thrips from each collection site were immediately preserved in 95% ethanol and returned to the laboratory at the University of California Riverside (UCR). All specimens were confirmed as matching the morphological description of C. fasciatus (Hoddle et al., Reference Hoddle, Mound and Paris2008) and stored at −20 °C. Whole genomic DNA was extracted from individual specimens using two methods. The majority of specimens were extracted using a simple Chelex method (Walsh et al., Reference Walsh, Metzger and Higuchi1991), in which individual thrips were first ground up in 3μl proteinase-K (>600mAUml−1; Qiagen, Valencia, CA, USA). To this was added 80μl of a 5% (w/v) suspension of Chelex® 100 resin (Bio-Rad Laboratories, Hercules, CA, USA) and the reaction was incubated at 55 °C for one hour followed by 10min at 99 °C. DNA was also extracted from specimens using the ‘salting-out’ technique described in Rugman-Jones et al. (Reference Rugman-Jones, Hoddle, Mound and Stouthamer2006) with the modification that specimens were ground up prior to incubation.
Table 1. Collecting details (from north to south) for Caliothrips fasciatus in California.

Amplification and sequencing
The polymerase chain reaction (PCR) was used to amplify a section of the mitochondrial gene (mtDNA) cytochrome oxidase c subunit 1 (COI) commonly used for DNA barcoding studies (Hebert et al., Reference Hebert, Cyminska, Ball and deWaard2003). PCR was performed in 25μl reactions containing 2μl of DNA template (concentration not determined), 1 × ThermoPol PCR Buffer (New England BioLabs, Ipswich, MA, USA), an additional 1mMMgCl2, 200μM each dNTP, 4% (v/v) BSA (NEB), 1U Taq polymerase (NEB) and 0.2μM each of the primers LCO1490 and HCO2198 (Folmer et al., Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994). Thermocycling was performed in a Mastercycler® ep gradient S thermocycler (Eppendorf North America Inc., New York, NY, USA) programmed for an initial denaturing step of 2min at 94 °C; followed by five cycles of 30s at 94 °C, 1min 30s at 45 °C and 1min at 72 °C; followed by a further 35 cycles of 30s at 94 °C, 1min 30s at 51 °C and 1min at 72 °C; and a final extension of 5min at 72 °C. Amplification was confirmed by gel electrophoresis and PCR products were subsequently cleaned using the Wizard® PCR Preps DNA purification system (Promega, Madison, WI, USA) and direct-sequenced in both directions at the UCR Genomics Institute, Core Instrumentation Facility.
Due to high levels of mitochondrial genetic diversity (see Results), we also amplified and sequenced a section of the conserved D2 domain of 28S rDNA (28SD2) from at least one specimen bearing each COI haplotype, using the primers 28sF3633 and 28sR4076 (Rugman-Jones et al., Reference Rugman-Jones, Hoddle and Stouthamer2010).
Sequence analysis
All sequences were trimmed and aligned manually in BioEdit 7.0.5.3 (Hall, Reference Hall1999). The alignment of COI sequences resulted in a matrix of 174 sequences, each 655bp in length. The absence of nuclear pseudogenes (Song et al., Reference Song, Buhay, Whiting and Crandall2008) was confirmed by translating the COI sequences using the Transeq application in EMBOSS (http://www.ebi.ac.uk/Tools/emboss/transeq/index.html), and representative sequences were subsequently deposited in GenBank® (accessions JQ609545–609600) (Benson et al., Reference Benson, Karsch-Mizrachi, Lipman, Ostell and Wheeler2008). Sequence polymorphism, within the dataset as a whole, was assessed as number of haplotypes, haplotype diversity (Hd), nucleotide diversity (π) and the average number of pairwise nucleotide differences (κ), all calculated using DnaSp Ver. 5.10.00 (Librado & Rozas, Reference Librado and Rozas2009). Aligned sequences of 28SD2 were identical (across all COI haplotypes) and were deposited in GenBank® (accessions JQ609601–609656), but not subjected to further analysis.
Relationships among the COI haplotypes were investigated using parsimony analyses (MP), conducted with TNT v.1.1 (Goloboff et al., Reference Goloboff, Farris and Nixon2003, Reference Goloboff, Farris and Nixon2008). Heuristic searches were performed using a New Technology Search with default settings, except for using a sectorial search, ratchet weighting probability of 5% with 50 iterations, tree-drifting of 50 cycles, tree-fusing of five rounds, and best score hit of 25 times, followed by swapping to completion on all trees found. Support was calculated using 1000 bootstrap replicates. The same trees were recovered using PAUP 4.0* (Swofford, Reference Swofford2002). Successive approximations character weighting (SAW) analysis was applied using the rescaled consistency index and a base weight of 1000 to analyze data stability (Babcock et al., Reference Babcock, Heraty, DeBarro, Driver and Schmidt2001). Maximum likelihood (ML) analyses were conducted with RAxML v.7.2.7 via the CIPRES Science Gateway (http://www.phylo.org). CIPRES was used to conduct rapid bootstrap analyses (RBS) (sensu Stamatakis et al., Reference Stamatakis, Hoover and Rougemount2008), where 1000 bootstrap analyses are conducted first, followed by fast and then slow searches on the sampled trees to find the best maximum likelihood tree. This revealed four groups of haplotypes (see Results; fig. 1) within and between which the average number of pairwise nucleotide differences (κ) was calculated. Average sequence divergence between the four groups was further quantified by calculating Kimura 2-P distances using MEGA version 5 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011).

Fig. 1. Phylogenetic relationships among COI haplotypes of Caliothrips fasciatus in California. best known likelihood tree from RAxML (−1574.594370). Alternate placement for group B2 resulting from parsimony analysis (P) indicated with arrow. Group B3 is monophyletic with 90% BS support in parsimony. Terminal taxa are numbered haplotypes. Bolded haplotypes (1, 27 and 32) are those used in the mating trials (see text) (![]() , Wolbachia + ve).
, Wolbachia + ve).
Estimates of genetic diversity (Hd, π, κ) were also examined within and between the Muir Woods (n = 34), Porterville (n = 28) and Riverside (n = 28) populations used as a source of the thrips haplotypes employed in the crossing experiments (see below). Pairwise estimates of F ST were obtained for these populations using the distance method implemented in ARLEQUIN ver. 3.11. (Excoffier et al., Reference Excoffier, Laval and Schneider2005). Significance of the F ST values was evaluated by permuting the haplotypes between populations, with the number of permutations set at 1000.
Mating trials
To facilitate inter-population mating trials, additional collections were made from three geographically separated locations along a north-south transect totaling 734km. The populations sampled were: (i) Muir Woods (Marin Co.), 37°53.587N 122°33.999W, 2 June 2008, from Eschscholzia californica Cham. (California poppy); (ii) Porterville (Tulare Co.), 36°03.858N 119°02.937W coordinates, 3 June 2008, from E. californica; and (iii) Riverside (Riverside Co.), 33°57.823N 117°20.312W, 8 June 2008, from Asparagus officinalis L. (asparagus). Live thrips were returned to the UCR Insectary and Quarantine Facility. Thirty females from each population were contained in individual double-ventilated vials containing a single Lima bean, Phaseolus lunatus L. leaf and a con-population male (for details of these vials see Irvin et al., Reference Irvin, Suarez-Espinoza and Hoddle2009). Females were allowed to mate and oviposit for several days, following which they were removed and their COI haplotype was determined. Only the offspring from those females that possessed the most common COI haplotype at each respective location were then combined to instigate the three mating trial populations. These populations were maintained in separate rearing rooms on caged, potted Lima beans (Var. Fordhook 242) at 26.6 °C ± 1.03 °C and 50% RH under long days (L:D 14:10h). Plants were watered every third day and fertilized with MiracleGro® (The Scotts Company LLC) at the recommended label rate. At 14-day intervals, new plants were introduced into cages, and existing plants were allowed to die, thereby forcing thrips onto new hosts. A strict management protocol was enforced to ensure colony cross contamination did not occur. This protocol involved showering and changing clothes before entering each colony. Voucher specimens from each of the three populations were deposited in the Entomology Research Museum at UCR (voucher numbers UCRC ENT 317104–UCRC ENT 317108).
All possible crosses were done between the three different source populations for a total of nine crosses. Each cross consisted of around 30 replicates of a male and a female isolated together in a double ventilated vial containing a single Lima bean leaf. Males and females were paired for four days to allow for mating, after which males were removed and females were left alone on the leaf for an additional seven days. Replicates were checked daily, and, if no larval development was seen after 14 days, the replicate was deemed an unsuccessful cross, and a new replicate initiated. This continued until approximately 30 successful pairings (i.e. pairings that produced offspring) were obtained for each cross. Thrips larvae produced from crosses were counted and reared to adulthood to allow determination of sex.
Virgin oviposition to assess thelytoky
To determine the sex of their offspring, 30 virgin females from each of the three populations used in the mating trials were isolated individually and allowed to oviposit on a bean leaf for 11 days as described above. However, these individually isolated virgin females unexpectedly failed to produce any offspring (see Results). The experiment was repeated, but, this time, virgin females were confined in groups of five. These conditions did result in offspring production (see Results); and, on reaching adulthood, the number of males and females was recorded.
Screening for Wolbachia infection
All females used in the mating trials, plus individuals randomly drawn from each of the remaining COI haplotypes, were screened for the presence of Wolbachia, an endosymbiont commonly associated with reproductive incompatibility and thelytoky in arthropods (Stouthamer et al., Reference Stouthamer, Breeuwer and Hurst1999). This was done using an assay based on PCR amplification of 16S rDNA with Wolbachia-specific primers (Werren & Windsor, Reference Werren and Windsor2000). PCR was performed in 25μl reactions containing 2μl of DNA template (concentration not determined), 1 × ThermoPol PCR Buffer (NEB), an additional 1mMMgCl2, 200μM each dNTP, 6% (v/v) BSA (NEB), 1U Taq polymerase (NEB) and 0.2μM each of the primers W-Specf and W-Specr (Werren & Windsor, Reference Werren and Windsor2000). Thermocycling conditions were: an initial denaturing step of 2min at 94 °C; followed by 42 cycles of 30s at 94 °C, 45s at 55 °C, and 1min 30s at 72 °C; and, a final extension of 10min at 72 °C. Trichogramma pretiosum Riley (Hymenoptera: Trichogrammatidae), known to be infected with Wolbachia, were used as positive controls. Amplification was checked via electrophoresis of 5μl of PCR product on 1% agarose gels stained with ethidium bromide, and specimens yielding a band of the appropriate size (438bp) were scored as positive for Wolbachia infection.
Statistical analysis of mating trial data
All analyses were performed in MINITAB® 15.1.30.0. (Minitab Inc.). We first investigated whether the genetic identity of a male influenced the likelihood of a successful mating with females from each of the three selected populations. Pairings that resulted in offspring were considered successful. Offspring production (‘yes’ or ‘no’) was used as a binary response variable in a logistic regression model with a logit link function. Male population origin was entered as the predicting factor, and the effect of hetero-population pairing (e.g. Muir Woods (M) female × Porterville (P) male) on successful mating was assessed in terms of any change in the likelihood of offspring production by examining the resulting regression coefficient and odds ratio relative to a con-population cross (e.g. M female × M male). For pairings that successfully produced offspring, the effect of male origin on the fecundity of females was investigated by comparing the total number of resulting offspring in a general linear model. Two fixed factors (female and male haplotype), each with three levels representing population origin (M, P and R (Riverside)), were entered into the model along with the interaction between the factors. Differences in the main effects were investigated with post-hoc Tukey's pairwise comparisons. Given their haplodiploid mechanism of sex determination, if hetero-population mating results in a decline in the total number of offspring relative to con-population mating (see Results); then, if this is caused by gametic incompatibility, this should be evident as a decline in the number of female offspring as a result of increased embryonic mortality due to fertilization problems. In contrast, the number of male offspring (which develop from unfertilized eggs) should not be affected. Thus, for pairings that successfully produced offspring, the effect of male origin on the production of male and female offspring was investigated by comparing the total number of each sex separately, using general linear models which were otherwise identical to that described for analyzing the total number of offspring (see above). Thus, in the event that reduced offspring production following a hetero-population mating is the result of gametic incompatibility, we expect females from each population to produce fewer female offspring when paired with a hetero-population male. This should be revealed in the general linear model for female offspring production (but not male production) as a significant interaction term between male and female population origins.
Results
Genetic variation
Among the COI sequences of 173 individuals collected across California, a total of 75 polymorphic sites and 56 haplotypes were observed (fig. 1). Haplotype diversity (Hd) was 0.937 ± 0.009 (mean ± SD), nucleotide diversity (π) was 0.020 ± 0.001 and the average number of nucleotide differences (κ) was 13.292. The two most divergent haplotypes, 43 and 52 (detected in the Mentone and Idyllwild populations respectively (table 1, fig. 1)), differed by 37bp (5.6%). All nucleotide substitutions were synonymous, with the exception of a single substitution at nucleotide position 368, present in three closely related haplotypes, each detected in only one specimen (haplotypes 54–56; fig. 1).
Eight equally parsimonious trees were found in the MP analysis (length 119, r.i. 0.63, c.i. 0.91; results not shown). Two trees of the same length resulted after SAW weighting, so the data were regarded as stable. The only conflict in the two trees (evident in both the parsimony and SAW results) was in the placement of haplotypes 38 and 39 + 40 within clade B3. The single tree resulting from the ML analysis (fig. 1) was similar to the MP results except for the placement of haplotypes 24–28 (group B2), which in the MP analysis instead formed a grade leading to monophyletic B1 and B3 groups (shift to P; fig. 1). The opposing likelihood hypothesis had very weak bootstrap support (53; fig. 1). Interestingly, although unresolved in the ML results, group B3 was strongly supported as monophyletic in the MP analyses with 90% bootstrap support. Taken together, MP and ML analyses suggested the haplotypes clustered into four groups (fig. 1). Average sequence divergence (Kimura 2-P distance) between these groups is shown in table 2.
Table 2. Genetic differentiation between the four groups of Caliothrips fasciatus haplotypes (revealed by MP and ML analyses; fig. 1) expressed as the average number of pairwise differences (κx,y) in a 655bp section of the COI gene (above the diagonal) and percent divergence (Kimura 2-P distances) (below the diagonal). Diagonal element is within-group κ.

Considering just the individual populations used in the mating trials, the least variable population was that from Muir Woods (n = 34). Here, the number of polymorphic sites was 21, the number of haplotypes was six, Hd = 0.369 ± 0.103, π = 0.007 ± 0.002 and κ = 4.549. The remaining two populations displayed slightly higher levels of variation. Within the Porterville population (n = 28), the number of polymorphic sites was 45, the number of haplotypes was ten, Hd = 0.791 ± 0.067, π = 0.009 ± 0.003 and κ = 6.116. Within the Riverside population (n = 28), the number of polymorphic sites was 42, the number of haplotypes was ten, Hd = 0.767 ± 0.065, π = 0.010 ± 0.004 and κ = 6.839. The distribution of haplotypes among these three populations was very structured with the northernmost and southernmost populations, Muir Woods and Riverside, respectively, sharing no haplotypes, and the geographically intermediate Porterville population sharing only two haplotypes with each of the other two populations. This was reflected in two pairwise measures of population differentiation κ and F ST (table 3). Of the three haplotypes chosen for use in the mating trials (fig. 1), the haplotypes of Porterville (1) and Riverside (32) were the most divergent (14bp or 2.21% difference) and the haplotype of Muir Woods (27) differed from each of these by 12bp (1.83%).
Table 3. Genetic differentiation between the three populations of Caliothrips fasciatus used in crossing experiments, shown as the average number of pairwise differences, κx,y (above the diagonal) in a 655bp section of the COI gene and as estimates of F ST (below the diagonal) calculated using the same gene region. The F ST estimates were all highly significant (P < 0.001) after sequential Bonferroni correction.

Population crosses
Crosses had the highest rate of success (i.e. they resulted in offspring production) when females were paired with males of their own population (tables 4 and 5). In females from each population, the odds that confinement with a hetero-population male would result in a successful mating (i.e. offspring production) were typically highly reduced (P < 0.001) at 8–16% of the odds for confinement with a con-population male (table 5). The only exception to this was in R females mated to P males where, although the odds appeared to be reduced to 49%, this was not statistically significant (table 5).
Table 4. Mating success (determined by the production of offspring) of reciprocal pairs of males and females drawn from three California populations of Caliothrips fasciatus.
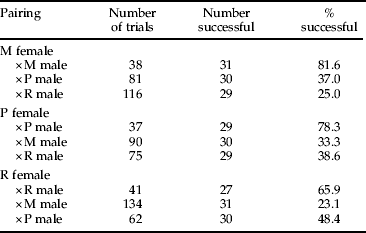
M, Muir Woods; P, Porterville; R, Riverside.
Table 5. Result of binary logistic regression analyses revealing reduced mating success (determined by the production of offspring) of hetero-population crosses relative to con-population crosses in Californian populations of Caliothrips fasciatus.
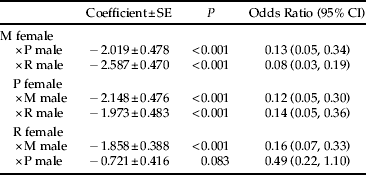
M, Muir Woods; P, Porterville; R, Riverside.
Similarly, following a successful cross, offspring production was always substantially higher when females mated with males from their own population as opposed to males of the other populations, revealed by a significant interaction between female and male population origins in the general linear model (F4,258 = 16.56, P < 0.001; fig. 2). Among the successful crosses, there were also significant main effects on fecundity caused by: female population origin (F2,258 = 8.17, P < 0.001), with R females producing more offspring than those from M (P = 0.006) and P (P < 0.001); and, male population origin (F2,258 = 7.28, P = 0.001), with R males inducing greater offspring production than P males (P < 0.001).

Fig. 2. Mean number of offspring produced by Caliothrips fasciatus females following successful con- or hetero-population matings. Error bars represent ± 1 standard error (![]() , Muir Woods; □, Porterville;
, Muir Woods; □, Porterville; ![]() , Riverside).
, Riverside).
The sex ratio of offspring from successful crosses was highly female biased, with the proportion of females ranging from 0.62–0.86 of offspring produced. When male and female offspring were considered separately, the number of male offspring produced was not influenced by female population origin (F2,258 = 0.51, P = 0.601), male population origin (F2,258 = 0.64, P = 0.528) or the interaction between male and female population origins (F4,258 = 0.87, P = 0.484) (fig. 3a). However, the number of female offspring produced was affected by female population origin (F2,258 = 9.55, P < 0.001; fig. 3b), with R females overall producing more female offspring than those from M (P = 0.007) and P (P < 0.001). Similarly, male population origin also influenced the number of female offspring (F2,258 = 7.32, P = 0.001; fig. 3b), with females paired with an R male producing more female offspring then those paired with a P male (P < 0.001). Finally, within each population, females produced the highest number of female offspring when paired with a con-population male, evident as a significant interaction between female and male population origins in the general linear model (F4,258 = 18.53, P < 0.001; fig. 3b).

Fig. 3. The effect of hetero-population mating on the number of (a) male and (b) female offspring. Error bars represent ± 1 standard error. Note difference in the scale of the y-axis ((a)![]() , Muir Woods; □, Porterville;
, Muir Woods; □, Porterville; ![]() , Riverside) (b)
, Riverside) (b)![]() , Muir Woods; □, Porterville;
, Muir Woods; □, Porterville; ![]() , Riverside).
, Riverside).
Virgin oviposition to assess thelytoky
In our initial assay, of the 90 virgin females (30 from each of the three experimental populations) confined individually for 11 days on a lima bean leaf, none produced any offspring. In contrast, when isolated as groups of five virgin females, 90% of the groups produced offspring. All offspring produced were male.
Wolbachia infection status
All C. fasciatus females used in the mating trials tested negative for infection with Wolbachia. Across our extended California sample, the majority of individuals (males and females) also tested negative. Exceptions to this were three male specimens: two from Santa Barbara Botanic Gardens and one from Carpinteria (both locations in Santa Barbara County, California). Interestingly, these were the same three specimens (and three haplotypes) that possessed the non-synonymous COI nucleotide substitution (fig. 1).
Discussion
In geographically isolated populations, adaptive genetic divergence may incidentally lead to the evolution of reproductive isolation and, hence, speciation (Muller, Reference Muller1942; Mayr, Reference Mayr1963). In populations that occupy different host plants and/or habitats, the rate at which such divergence occurs is likely to accelerate as the populations actively adapt to differing environmental conditions (Funk, Reference Funk1998; Nosil, Reference Nosil2007). Processes similar to these may be acting on populations of C. fasciatus. We examined variation across Californian populations of this thrips and found evidence for genetic divergence and outbreeding depression between allopatric populations.
Sequences of the COI gene of mtDNA revealed the existence of a large number of haplotypes (56), and these separated into two major groups with 100% bootstrap support (fig. 1). Group A consisted of two clades: one found only near Santa Barbara (haplotypes 54–56), and a second clade with a wider distribution in southern California (Riverside, San Bernardino and Kern Counties, haplotypes 51–53). Haplotypes in group B split into three subgroups with lower bootstrap support: one group (B1; haplotypes 1–23) with a mainly northern distribution stretching from the southern end of the San Joaquin Valley into northern California, and two more closely related groups (B2 and B3, haplotypes 24–28 and 29–50, respectively) with a more southern distribution (haplotypes 24–50). Although this general pattern was strong, the separation into geographic localities was not perfect. For example, haplotype 27 grouped with the ‘southern’ haplotypes but was only collected in Muir Woods (northern California); and, in several other locations, ‘southern’ and ‘northern’ haplotypes mixed (see table 1). The maximum sequence divergence between the four groups (A, B1, B2 and B3) is given in table 2. Group A differs most from the other groups, and the average K2-P distances between its members and members of each of the B subgroups (3.63–4.80%) may indicate that group A represents a different species from group B.
In the crossing experiments, we chose three populations (all from the B group) from different geographical areas in California to determine their crossing compatibility. To construct the populations for the crossing experiments, for each population, we chose individuals with a particular COI haplotype. Since at the time of setting up this assay we did not know if cryptic species might be present, using only individuals with a single haplotype for each population reduced the chance that any population would consist of a mixture of cryptic species, which would influence the outcome of crosses using each population. The presence of cryptic species within a population has previously been shown in other thrips (Rugman-Jones et al., Reference Rugman-Jones, Hoddle and Stouthamer2010). For the Muir Woods population, we chose individuals with haplotype 27 (group B2); for Porterville, haplotype 1 (group B1); and, for Riverside, haplotype 32 (group B3). If these populations all belong to the same species, then the haplotype should be a neutral factor in the outcome of our crosses, and the results of the crosses should only be affected by the nuclear genes. Potential effects of symbionts on mating compatibility were excluded by testing C. fasciatus for Wolbachia infection. In addition, in preliminary analyses of thrips populations (data not shown), we also found no evidence for the presence of another symbiont Cardinium, which may also influence crossing compatibility between infected and uninfected individuals (Hunter et al., Reference Hunter, Perlman and Kelly2003).
Our crossing studies show that females paired with males from their own population were the most successful, both in the fraction of all pairings that successfully resulted in offspring, and in total number of offspring produced by those successful pairs. Following a successful mating, females of a particular population always produced similar numbers of male offspring, regardless of male population origin. This is consistent with the assumption that females fertilize the same proportion of their eggs independent of the origin of their mate. However, the significant reduction in the number of female offspring produced when females mated with hetero-population males indicated a level of post-zygotic incompatibility between the two populations. In addition, a level of pre-zygotic incompatibility also appears to exist, as indicated by the significantly lower success rate of hetero-population pairings compared to con-population pairings. However, since we did not observe mating pairs, we do not know if reduced success rates were due to a lack of mating or if mating took place but did not result in oviposition by the females. The former possibility appears the most likely since we also found that isolated virgin females did not produce any offspring. Despite strong evidence for outbreeding depression in our crossing study, it is obvious that the different populations are somewhat compatible and reflect the same species. Kimura 2 parameter distances (1.88–2.64) between the B1, B2 and B3 haplotypes (fig. 1), also suggest that these groups constitute a single species (Hebert et al., Reference Hebert, Cyminska, Ball and deWaard2003). This is further supported by the lack of variation in a fragment of a second gene that was sequenced, the ribosomal 28S. While 28S is highly conserved and little or no variation is expected within a species, closely related species can sometimes be detected by slight differences (see Rugman-Jones et al., Reference Rugman-Jones, Hoddle and Stouthamer2010).
The female biased offspring sex ratios resulting from our crosses are difficult to explain. Unless there is elevated mortality of male versus female eggs, and/or male and female offspring require different levels of maternal investment, theory would predict a balanced sex ratio of 50% males and 50% females. Local mate competition, which is often the cause of female biased sex ratios in sib-mating parasitic Hymenoptera, seems unlikely in these thrips since their oviposition pattern should result in random mating between males and females in a population. The offspring sex ratio found in our study is very similar to the field population sex ratios found by Bailey (Reference Bailey1933) that ranged from a low of 57% females, to a high of 74% females, with an average sex ratio over a year of 66.3% females. Female biased laboratory population sex ratios, with no evidence for differential mortality of males and females, have also been reported for Ceratothripoides claratris (Shumsha) (Thysanoptera: Thripidae), but only when reared at 30 °C or higher (Premachandra et al., Reference Premachandra, Borgemeister, Chabi-Olaye and Poehling2004). Female-biased field population sex ratios have also been reported in other thrips species (Vasiliu-Oromulu, Reference Vasiliu-Oromulu, Marullo and Mound2002; Šmatas, Reference Šmatas2009), but factors underlying these ratios are rarely studied (Crespi, Reference Crespi, Wrensch and Ebbert1993).
The lack of offspring production by individual virgin female bean thrips held in isolation was unexpected since thrips are haplodiploid and can consequently produce sons from unfertilized eggs. The lack of offspring production by isolated virgin females is not a general feature of this order. Kumm & Moritz (Reference Kumm and Moritz2010) show that isolated F. occidentalis virgin females readily produce sons (and, in rare cases, even the occasional daughter) and similar results have been observed for C. claratris (Premachandra et al., Reference Premachandra, Borgemeister, Chabi-Olaye and Poehling2004), Franklinothrips orizabensis Johansen (Thysanoptera: Aeolothripidae) (Hoddle et al., Reference Hoddle, Robinson, Drescher and Jones2000) and Scirtothrips perseae Nakahara (Thysanoptera: Thripidae) (Hoddle, Reference Hoddle2002). Furthermore, in his original work on C. fasciatus, Bailey (Reference Bailey1933) reported that two individual virgin females produced male offspring. Bailey's (Reference Bailey1933) experiments differed from ours in that he gave virgin females 27 days to produce offspring, while we ended our experiments with isolated virgin females after 11 days. Thus, female age may influence virgin oviposition. Although we tried unsuccessfully to get individual virgin females to produce offspring, when we isolated groups of five virgin females together, the majority of groups (90%) produced male offspring, with oviposition taking place as early as day four post-grouping. Thus, it seems that the presence of other females may stimulate oviposition in virgin females, although the underlying reason for this is unknown. One possibility is that females ‘hedge their bets’. There is evidence that insects alter their reproductive/oviposition strategies in a variety of ways in response to their immediate environment (Gage & Baker, Reference Gage and Baker1991; Hopper, Reference Hopper1999). If a female is not mated and there are also no other conspecific females around (i.e. an isolated virgin), it may be advantageous for her to defer oviposition, since any sons she produces would have a low chance of encountering a conspecific female with which to mate (perhaps because of limited dispersal ability). There is probably a tradeoff between oviposition and longevity, and as she ages, the production of sons (despite their limited chance of encountering females) may be a mother's only chance of gaining any fitness at all. Reaching this tradeoff point could explain the difference between our findings and those of Bailey (Reference Bailey1933), who maintained his females for a longer period. In contrast, if other females are present (i.e. as in our groups of five virgins), although an individual female hasn't mated, she lacks information on the mating status of the others, which therefore may produce female offspring. Thus, unmated females may gain fitness by producing sons, which have a high chance of encountering a conspecific female. Indeed, under this scenario, since mated C. fasciatus females bias their offspring sex ratio towards daughters (see above), it may in fact be particularly beneficial for the virgin female to oviposit, because in fitness terms, sons would be more valuable than daughters (Godfray & Hardy, Reference Godfray, Hardy, Wrensch and Ebbert1993).
All COI sequences translated to the same protein with the exception of haplotypes 54–56 (group A), which carried a single non-synonymous change. These three haplotypes were each found in only one specimen, and those three specimens together were also the only ones infected with the bacterial symbiont Wolbachia, a bacterium known to cause several reproductive anomalies in insects such as parthenogenesis induction, cytoplasmic incompatibility, feminization and male-killing (Stouthamer et al., Reference Stouthamer, Breeuwer and Hurst1999). The three infected specimens were all males, making it unlikely that this Wolbachia infection results in parthenogenesis or in the male-killing phenotype. We do not know if this infection causes another reproductive effect, but it is clear that the infection is co-inherited with the mitochondrial type as expected. If the acquisition of Wolbachia was recent, it would be expected that only some individuals of a single haplotype will be infected. Therefore, the fact that three closely related haplotypes are infected could indicate that this is not a recent infection.
The three Wolbachia-infected haplotypes (54–56) were all collected from the Santa Barbara area amidst uninfected specimens of other haplotypes (table 1). The Wolbachia-infected COI haplotypes were most closely related to another group of three haplotypes (51–53) detected in specimens from inland areas of southern California; and, together, this group of six haplotypes (group A; fig. 1) was relatively divergent from the remaining 50 haplotypes. Indeed, average sequence divergence, based on Kimura two parameter distances (ranging from 3.63–4.80), suggests that these haplotypes may, in fact, represent a different species (Hebert et al., Reference Hebert, Cyminska, Ball and deWaard2003). However, confirming this would require additional genetic work to determine if there are also consistent nuclear genetic differences between specimens carrying group A and group B haplotypes. Reciprocal crossing experiments may also be used to reveal that A and B are cryptic species (or simply more divergent forms of C. fasciatus), but may be confounded if the Wolbachia found in haplotypes 54–56 causes any kind of crossing incompatibility. If that is the case, then it is also possible that the Wolbachia infection could spread rapidly through California populations of C. fasciatus, as happened in Drosophila simulans Sturtevant (Turelli & Hoffmann, Reference Turelli and Hoffmann1991).
In conclusion, bean thrips in California belonging to groups B1, B2 and B3 (fig. 1) appear to be a single species although significant pre- and post-zygotic isolation exists between geographically distant populations. We did not attempt to cross specimens with group A haplotypes with those possessing group B haplotypes and, therefore, are not sure if group A constitutes a cryptic species or is simply a more divergent form of C. fasciatus. In general, California bean thrips (belonging to group B) are not infected with the bacterial symbiont Wolbachia; however, within the C. fasciatus population belonging to group A, one of the two subclades is infected with Wolbachia. The phenotype associated with this infection is unclear but thelytoky and male killing are unlikely.
Consequences for the invasion potential of C. fasciatus outside its native range
Caliothrips fasciatus has failed to establish in areas outside of its native range despite being exported from California inside the navels of oranges since at least 1899 (Hoddle et al., Reference Hoddle, Stosic and Mound2006; Mound et al., Reference Mound, Zhang and Bei2011). Since the 1960s, among the substantial volumes of fruit exported from California to countries that have had suitable habitat and climate for establishment, a small percentage of navel oranges has regularly been contaminated with C. fasciatus (Hoddle et al., Reference Hoddle, Stosic and Mound2006). As a result, this native California pest has a high quarantine rating in several receiving countries (e.g. Australia Phyto Requirements, 2010). Despite this persistent propagule pressure in areas that are favorable for establishment, C. fasciatus has not successfully invaded (Hoddle et al., Reference Hoddle, Stosic and Mound2006). The results of this study suggest several factors may adversely affect the invasion potential of C. fasciatus. Factors identified here are: the absence of thelytoky, reducing the chances of a virgin female initiating a population; the reluctance of virgin females to produce male offspring, reducing the potential for mother-son matings and, hence, further reducing the chances of a virgin female initiating a population; and, reduced mating success between members of different populations which could occur if contaminated oranges from different areas of California were amalgamated in packinghouses and exported together. These factors, acting alone or in concert, may explain why C. fasciatus has been unable to establish outside of its native range from small founding populations accidentally exported in Californian navel oranges.
Acknowledgements
Christina Hoddle (UCR, who helped with field collections and the establishment of live specimens on bean plants in the field prior to transportation to UCR); Phil Phillips (UCCE Ventura County, collections for DNA); and, David Headrick (Cal Poly San Luis Obispo, collections for DNA).










