Introduction
Studies of resource use in insects have revealed much about the interactions between ecological and evolutionary forces, and there are now many excellent examples of the formation of cryptic species, ‘biotypes’ and ‘host races’ among populations of phytophagous insects feeding on different host plant species (Dres & Mallet, Reference Dres and Mallet2002; Funk et al., Reference Funk, Filchak and Feder2002). While these plant parasites have been well studied, the process of ecological diversification in other parasitic organisms has received comparatively little attention (Huyse et al., Reference Huyse, Pulin and Theron2005). Specifically, it remains unclear to what extent host plant associated differentiation and subsequent speciation in phytophagous insects triggers co-divergence in natural enemies, such as predators and parasitoids (Cronin & Abrahamson, Reference Cronin and Abrahamson2001; Stireman et al., Reference Stireman, Nason, Heard and Seehawer2006; Abrahamson & Blair, Reference Abrahamson, Blair and Tilmon2007). Given the increasing interest in coevolutionary processes, studies examining the degree to which higher trophic levels evolve in response to genetic structure of hosts or prey (and vice versa) will be important for understanding species interactions and diversification (Singer & Stireman, Reference Singer and Stireman2005), as well as for applied sciences such as biological control (Roderick & Navajas, Reference Roderick and Navajas2003; Hufbauer & Roderick, Reference Hufbauer and Roderick2005).
Parasitoids are organisms that lay their eggs in or on the body of an arthropod host, which is consumed and killed by the developing parasitoid larvae (reviewed by Godfray, Reference Godfray1994). The Aphidiinae (Hymenoptera: Braconidae) consists of ~400 species that are solitary endoparasitoids of aphids (Homoptera: Aphididae: Starý, Reference Starý, Minks and Harrewijn1988). Host range within the subfamily varies from strict specialization to broad generalism (Starý, Reference Starý, Minks and Harrewijn1988), though it is believed that many supposedly generalist taxa comprise cryptic host specific complexes (e.g. Tremblay & Pennacchio, Reference Tremblay, Pennacchio and Gupta1988; Atanassova et al., Reference Atanassova, Brookes, Loxdale and Powell1998). However, while host specificity has been well-studied in these parasitoids, there is little data on how often such specialization has evolved in concert with plant-associated diversification in their aphid hosts.
Aphidius transcaspicus Telenga is an aphidiine parasitoid that is distributed across the Mediterranean basin and central Asia. In the field, the species appears highly specific to aphids in the genus Hyalopterus Koch (Kavallieratos et al., Reference Kavallieratos, Tomanović, Starý, Athanassiou, Sarlis, Petrović, Niketić and Veroniki2004; personal communication), though it will parasitize other aphids with varying degrees of success in laboratory settings (Wang & Messing, Reference Wang and Messing2006; personal observation). Hyalopterus species have complex life-cycles involving cyclical parthenogenesis and host alternation between primary host plants in the genus Prunus, on which sexual reproduction and overwintering occurs, and a secondary host plant (typically Phragmites australis), where populations persist clonally during the summer months. Hyalopterus comprises three broadly sympatric species that diverged several million years before present and exhibit strong associations with three principal primary host plants (Lozier et al., Reference Lozier, Roderick and Mills2007, Reference Lozier, Foottit, Miller, Mills and Roderick2008) – H. amygdali with P. dulcis (almond), H. persikonus with P. persica (peach), and H. pruni with P. domestica (plum). These species have maintained their genetic isolation and associations with these host plants despite sharing a secondary host plant and frequent co-occurrence on the primary host plant P. armeniaca (apricot) and other less commonly utilized Prunus spp. (Lozier et al., Reference Lozier, Roderick and Mills2007). Aphidius transcaspicus attacks Hyalopterus on each of their primary and secondary host plants throughout the Mediterranean, though at present no ecological data are available regarding host preferences for the different Hyalopterus species or frequency of parasitism on different primary and secondary host plant species.
Given the host plant associated genetic differentiation documented in Hyalopterus and apparent specificity of A. transcaspicus to this aphid genus in the field (Kavallieratos et al., Reference Kavallieratos, Tomanović, Starý, Athanassiou, Sarlis, Petrović, Niketić and Veroniki2004), in this study we aimed to investigate whether host plant associations have helped structure genetic diversity across trophic levels, or whether other factors, such as geographic separation, may be more important for population differentiation in this parasitoid. Beyond the implications that a better understanding of diversification at different trophic levels holds for ecology and evolution, the identification of cryptic host plant associated structure in A. transcaspicus also has important practical significance. Hyalopterus pruni is the only member of Hyalopterus detected in North America (Lozier et al., in press) and is an important agricultural pest of dried plum in California. The discovery of parasitoid species or biotypes that are strongly associated with Hyalopterus on plum trees could be crucial for selecting the most specific and effective populations of A. transcaspicus for use in biological control (Gordh & Beardsley, Reference Gordh, Beardsley, Bellows and Fisher1999).
The evolution of host or host plant-specific lineages in a parasitoid, as for other insects, would require genetic polymorphism for traits associated with host use combined with barriers to gene flow and/or disruptive selection (reviewed in Via, Reference Via2001; Rundle & Nosil, Reference Rundle and Nosil2005; Abrahamson & Blair, Reference Abrahamson, Blair and Tilmon2007). Broadly distributed parasitoids might exhibit host specialization because of historical vicariance and geographic heterogeneity in the distribution of host species followed by subsequent expansion into sympatry (Althoff & Thompson, Reference Althoff and Thompson2001; Bush & Butlin, Reference Bush, Butlin, Dieckmann, Doebeli, Metz and Tautz2004; Vos & Vet, Reference Vos and Vet2004). Alternatively, differentiation might occur via host shifts in the absence of physical isolation when selection associated with hosts or host plants is strong (Rundle & Nosil, Reference Rundle and Nosil2005). The opportunity for ecologically mediated isolation may be high in aphidiine parasitoids due to a number of factors linked to their unique ecology. Importantly, host associated differentiation will likely depend both on the aphid host, which acts as a resource and a site for reproduction through oviposition, and the host plant, which provides the context in which this interaction occurs and can in some cases affect host preferences more strongly than the host itself (Storeck et al., Reference Storeck, Poppy, van Emden and Powell2000; Daza-Bustamante et al., Reference Daza-Bustamante, Fuentes-Contreras, Rodriguez, Figueroa and Niemeyer2002). It has been demonstrated in several aphidiines that chemical information obtained at adult emergence govern the host plant upon which mating and host foraging occurs (Vet & Dicke, Reference Vet and Dicke1992; Storeck et al., Reference Storeck, Poppy, van Emden and Powell2000), promoting correlations between preferences for mating, foraging and oviposition habitats. The tendency for siblings to mate at the emergence site is also common in many parasitoids (Godfray & Cook, Reference Godfray, Cook, Choe and Crespi1997; Mackauer & Volkl, Reference Mackauer and Volkl2002) and could enhance such pre-zygotic isolation by further restricting gene flow, increasing linkage disequilibrium among genes involved in habitat and mate preference or providing mating opportunities for isolated populations during the initial period following a host shift (Askew, Reference Askew1968; Via, Reference Via2001; Dieckmann & Doebeli, Reference Diekmann, Doebeli, Dieckmann, Doebeli, Metz and Tautz2004). Lastly, selection acting to increase the efficiency with which endoparasitoids respond to immune defenses or secondary symbiont communities of different host species, or to chemical differences associated with different plant species, may establish performance trade offs that can limit genetic exchange among populations (Tremblay & Pennacchio, Reference Tremblay, Pennacchio and Gupta1988; Henter, Reference Henter1995; Oliver et al., Reference Oliver, Russel, Moran and Hunter2005; but see Hufbauer, Reference Hufbauer2001).
Detecting cryptic lineages can be difficult in parasitoids due to a lack of good morphological characters, and identifying diversity in the genus Aphidius has been problematic even at the species level, with A. transcaspicus itself only recently validated as a distinct species (Kavallieratos & Lykouressis, Reference Kavallieratos and Lykouressis1999). However, molecular markers have proven useful at distinguishing cryptic diversity in parasitoids at the species and population level (e.g., Hoy et al., Reference Hoy, Jeyaprakash, Morakote, Lo and Nguyen2000; Smith et al., Reference Smith, Wood, Janzen, Hallwachs and Hebert2007). Thus, to assess potential cryptic differentiation in A. transcaspicus, we examined eight molecular markers including the mitochondrial (mtDNA) cytochrome oxidase I (COI) gene and seven microsatellites (Lozier et al., Reference Lozier, Mills and Roderick2006). This combination of markers is likely to provide insights into evolutionary processes at different timescales. Genealogical relationships among COI sequences can be useful for detecting differences among putative insect species (e.g. Smith et al., Reference Smith, Wood, Janzen, Hallwachs and Hebert2007) or well-resolved biotypes (e.g. Boykin et al., Reference Boykin, Shatters, Rosell, McKenzie, Bagnall, De Barro and Frohlich2007). Patterns of similarity among microsatellite genotypes, while of limited use for phylogenetics, can still provide support for such higher level differences at multiple independent loci and can also be used to infer relationships at finer scales between populations and individuals (Schlötterer & Pemberton, Reference Schlötterer, Pemberton, DeSalle and Schierwater1998). We present genetic data for parasitoids reared from Hyalopterus spp. collected on the four principal primary host plants (almond, apricot, peach and plum) from Spain and Greece to compare the relative importance of Prunus host plant species and spatial separation on patterns of genetic structure. Studies of genetic variation in host-parasite systems can reveal any of a number of potential evolutionary histories with varying degrees of congruence between patterns found in hosts and parasites (Banks & Paterson, Reference Banks and Paterson2005). At one extreme is the possibility that A. transcaspicus will show even greater affiliation to Prunus species than does Hyalopterus, which would be supported by well-resolved parasitoid lineages or genotypic clusters associated with each of the four host plants, irrespective of sampling location. Similarly, we might observe a pattern closer to that found in Hyalopterus (i.e. with geographically widespread plum, almond and peach associated lineages but no lineage specific to apricot) that would be consistent with co-cladogenesis of parasitoids with their aphid hosts but would not necessarily demonstrate affiliations to particular host plants. Alternatively, we might find a generalist A. transcaspicus that randomly mates with respect to host plants but is strongly differentiated among geographic regions. Such a pattern would be expected for a widespread parasitoid that has failed to diverge with its host and where geographically limited dispersal (i.e. isolation by distance) or allopatric barriers lead to differentiation among regions. Finally, at the opposite extreme, if A. transcaspicus is highly mobile and generalist with respect to host and host plant preferences, we may see little genetic differentiation at any scale.
Materials and methods
Sampling
Aphidius transcaspicus was collected from almond, apricot, peach and plum trees from 20 locations in Spain and Greece (here, we treat each sampling event as a distinct ‘population’; fig. 1). Parasitoids were collected as mummified aphids (hollowed out aphid husks representing the cocoon of the developing parasitoid) and reared individually to emergence before being transferred to 95% ethanol. Parasitoid collections were made at the same time as the Hyalopterus collections previously reported (Lozier et al., Reference Lozier, Roderick and Mills2007). While identification of the Hyalopterus species from which individual A. transcaspicus emerged would have been difficult if not impossible, based on our previous results it is highly likely that parasitoids collected on almond, peach and plum would be from H. amygdali, H. persikonus and H. pruni, respectively; and parasitoids from apricot could come from any of these Hyalopterus species. We note that this approach is similar to that used in our previous study of Hyalopterus (Lozier et al., Reference Lozier, Roderick and Mills2007), where we used no prior taxonomic knowledge in our inference of genetic relationships and which proved effective for distinguishing the overall signal of host associated differentiation as well as more complex patterns of population admixture. However, we do consider the implications of occasional ‘wrong’ host plant choices by Hyalopterus on the genetic structure of A. transcaspicus (see Discussion). Lastly, we note that parasitoid mummies were encountered most frequently on almond and were unfortunately less prevalent on other Prunus spp., though we were able to include at least one locality for each plant species in both regions.

Fig. 1. (a) Regions sampled for Aphidius transcaspicus with detailed insets for collecting localities in (b) Spain and (c) Greece. Number of samples genotyped in Spain, June 2002 (b): S14almond=10, S15plum=10, S22peach=14, S23apricot=9, S24apricot=10, S25plum=12, S26almond=4, S28almond=10, S31almond=9; for Greece, May 2003 (c): Ga20almond=10; Ga06almond=8, Ga08almond=6, Ga12almond=12; for Greece, May 2005 (c): Gb04plum=13, Gb05almond=12, Gb06peach=12, Gb09apricot=10, Gb10almond=12, Gb12plum=2, Gb14peach=5 (![]() , almond; +, apricot; ▲, peach;
, almond; +, apricot; ▲, peach; ![]() , plum).
, plum).
DNA sequencing
Genomic DNA was extracted from whole female parasitoids using a Qiagen DNEasy DNA extraction kit (Qiagen Corporation, California, USA), eluted into 100–150 μl, and stored at −20°C. We amplified COI using the primers C1-J-1718 and C1-N-2191 (Simon et al., Reference Simon, Frati, Beckenbach, Crespi, Liu and Flook1994). All COI polymerase chain reaction (PCR) amplifications used for sequencing were performed in 10 μl volumes as in Lozier et al. (Reference Lozier, Roderick and Mills2007: except with an annealing temperature of 52°C) and products were purified using ExoSAP-IT (USB Corporation, Ohio, USA). Both PCR strands were cycle-sequenced in 10 μl volumes using the same primers under the following cycle sequencing conditions: 0.6 μl BigDye v3.1 (ABI), 0.5 μl 5× sequencing buffer, 4.0–5.0 pmol primer, and 1.2 μl purified PCR product. Sequencing products were purified using Sephadex (Sigma-Aldrich, Missouri, USA) and electrophoresis was performed on an ABI 3730. Sequences were aligned in Sequencher 4.0 (Gene Codes Corporation, Michigan, USA) and deposited in GENBANK with the accession numbers EF541030–EF541108.
Microsatellite genotyping
We genotyped 190 A. transcaspicus females at nine microsatellite loci (At001, At003, At004, At005, At006, At009, At014, At016 and At017), performing PCR amplification, +A overhang removal and electrophoresis on an ABI 3730 as in Lozier et al. (Reference Lozier, Mills and Roderick2006, Reference Lozier, Roderick and Mills2007). To minimize laboratory error, all reaction steps were performed simultaneously with multiple positive (known genotypes) and negative (sterile water) controls, and the qualities of allele size calls were checked manually. Any genotypes that could not be reliably scored were reamplified and, if necessary, were excluded from the study. Of the 1710 total genotypes attempted, we were unable to amplify only 15.
COI analyses
We examined the distribution of COI diversity by constructing a 95% confidence statistical parsimony network using TCS (Clement et al., Reference Clement, Posada and Crandall2000). The network was visualized with sequences pooled either by host plant or geographic origin. We tested the significance of the association between haplotypes and host plant or geography using contingency table analysis with the Pearson X2 statistic (the test is only performed for haplotypes A and B to avoid problems of low expected cell counts due to infrequent haplotypes; see below). We further tested population structure by examining the genetic covariance ‘Among Groups’ (F CT), ‘Among Populations within Groups’ (F SC), and ‘Among Individuals within Populations’ (F ST) using analysis of molecular variance (AMOVA) implemented in ARLEQUIN 3.1 (Excoffier et al., Reference Excoffier, Laval and Schneider2005). We tested the significance of two population groupings, (i) by geographic region and (ii) by host plant. Significance of each statistic was tested by 1000 permutations of the appropriate hierarchical units (table 1). To examine the possible confounding effect of apricot, which is a host plant shared by all three Hyalopterus species, we tested the host plant group hypothesis both with and without parasitoids from this host plant.
Table 1. AMOVAs of Aphidius transcaspicus COI sequence data grouped by (a) geographic region and by (b) host plant/host plant without apricot samples.
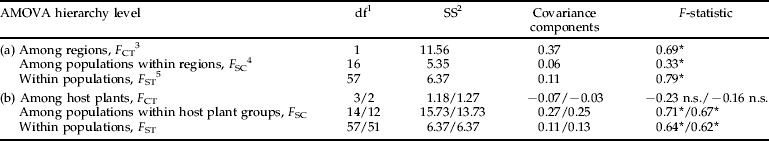
1 Degrees of freedom, 2 Sum of squares, 3F ST (and within population covariance component) is tested by permutation of genotypes among populations and groups, 4F SC (and among population within groups covariance component) is tested by permutation of genotypes among populations within groups and 5F CT (and among groups covariance component) is tested by permutation of populations among groups.
* P<0.001; n.s., not significant.
Basic microsatellite analyses
For the microsatellite data, we used FSTAT (Goudet, Reference Goudet2001) to calculate locus-specific diversity (Nei & Kumar, Reference Nei and Kumar2000) and F (Weir & Cockerham, Reference Weir and Cockerham1984) statistics and to test for deviations from Hardy-Weinberg Equilibrium (HWE) using 3600 permutations of alleles among individuals and F IS as a test statistic (table 2). We used GENEPOP (Raymond & Rousset, Reference Raymond and Rousset1995) to test for linkage disequilibrium (LD) with Fisher's method, correcting for the large number of tests with the Bonferroni method. Population structure was assessed using pairwise estimates of F ST, with significance for each pair tested by randomizing multilocus genotypes among populations 1000 times and correlations with geographic distance examined using Mantel tests (5000 permutations) in FSTAT. We also used partial Mantel tests to examine the significance of host plant effects on this correlation, coding population pairs from the same plant species as 1, and from different species as 0 (a negative correlation coefficient, r, would thus indicate that population pairs from different host plants were more greatly differentiated than from the same plant species). We then used AMOVA to test the significance of partitioning genetic variance among regions and host plant groups as described above, also examining structure separately within Spain and Greece.
Table 2. Locus specific diversity statistics for the entire sample of Aphidius transcaspicus from Spain and Greece.

1 Observed heterozygosity, 2 within population heterozygosity, 3 total heterozygosity (Nei & Kumar, Reference Nei and Kumar2000) and 4 correlation of alleles (F IT), correlation of alleles within populations (F ST), and correlation of alleles within individuals within populations (F IS) (Weir & Cockerham, Reference Weir and Cockerham1984) and their standard errors (estimated by jackknifing over populations using FSTAT).
*** Significant deviation from within-population Hardy-Weinberg Equilibrium, P<0.001.
Clustering analyses
Standard genetic analyses like AMOVA test differences among population groups specified a priori. Thus, we employed an alternative model-based analysis using the program BAPS 5.0 (Corander & Marttinen, Reference Corander and Marttinen2006), which implements a Bayesian mixture model to infer genetic clusters based solely on patterns of microsatellite variation and not on any prior grouping of populations. Patterns of clustering from such methods have proven useful for detecting cryptic host associated genetic structure in natural populations (e.g. Lozier et al., Reference Lozier, Roderick and Mills2007) and would provide evidence for host associated differentiation if assignment to clusters aligned with plant species, either across the entire sampled area (e.g. for cryptic species) or independently within either geographic region (e.g. for local differentiation). In contrast, assignment of populations from different plant species to the same cluster could be inferred as evidence against host associated differentiation. To determine optimum population partitioning, BAPS was first run using the ‘Clustering Groups of Individuals’ option. The number of potential clusters (K) was set from 1 to 20, with each specified K tested three times per run and results merged to obtain the optimum number of partitions based on likelihood scores. To avoid biasing results towards geographic or host associated clustering, we did not specify locality coordinates. The resulting clusters were then used in the admixture analysis to obtain posterior estimates of the proportion of individual genotypes represented by each cluster (admixture coefficients) using the following program settings: (i) minimum number of individuals=5; (ii) number of iterations used to estimate admixture coefficients=100; (iii) number of reference individuals=200; (iv) number of iterations used to estimate admixture coefficients for reference individuals=20 (see BAPS manual; http://web.abo.fi/fak/mnf//mate/jc/software/).
Lastly, we examined relationships among individuals using a tree-based clustering analysis of the microsatellite data. We used the software MICROSAT 2 (Minch, Reference Minch1997) to calculate the Cavalli-Sforza chord distance (D C: Cavalli-Sforza & Edwards, Reference Cavalli-Sforza and Edwards1967) among individuals and constructed an unrooted neighbor-joining (NJ) tree from observed distances using PHYLIP v3.67 (Felsenstein, Reference Felsenstein2004). We assessed tree support using a majority rule consensus NJ tree from 1500 bootstrap replicates (performed in MICROSAT 2) in PHYLIP, recording those clades found in more than 50% of the bootstrap replicates.
Results
COI sequence data
We identified only four 428 bp haplotypes in all A. transcaspicus individuals sequenced at COI. We limited our sequence analysis to a subset of 75 (out of 190 total) individuals once this low diversity became apparent in preliminary analyses, as it was deemed unlikely that increased sequencing would reveal additional structure indicative of host associated species. Haplotype A was the most frequently recovered sequence (n=41), followed in order by B (n=28), C (n=4) and D (n=2). There is no apparent pattern of host plant associations, with the two most common haplotypes present on all four Prunus species (X2=2.3, n=69; P=0.51; fig. 2a). The association is also insignificant when apricot is excluded from the analysis (X2=2.057, n=60, P=0.36). The geographic pattern is significant, however, with all four haplotypes present in Greece (+Crete), but parasitoids from Spain possessing only haplotype B (X2=44.2, n=69, P<0.001; fig. 2b). The AMOVA supports these patterns, with highly significant and substantial ‘Among Geographic Region’ group structure (F CT=0.69, P<0.001; table 1a) but an absence of significant ‘Among Host Plant’ group structure (F CT=−0.23, P=0.97; table 1b). If anything, the negative covariance and F CT for ‘Among Host Plant’ group structure indicates greater variance among populations from the same plant species than among populations from different plant species, though it is probably more conservative to interpret these values as a lack of differentiation. However, it seems likely that the large covariance component for the ‘Among Populations within Host Plant’ group level (table 1b) can be attributed to the strong differentiation among populations sampled from the same plant species but in different regions. Tests performed without samples from apricot gave similar results (table 1b–d).

Fig. 2. COI haplotype network for Aphidius transcaspicus, with circles and edges representing unique haplotypes and single base pair mutations, respectively. Circle areas are scaled approximately by the numbers of samples that possessed a given haplotype (75 total samples), with shading representing the proportion of samples associated with (a) host plant and (b) geographical location. Sample sizes: Spain-almond: 7; Spain-apricot: 3; Spain-peach: 6; Spain-plum: 5; Greece-almond: 2; Crete-almond: 23; Crete-apricot: 6; Crete-peach: 13; Crete-plum: 11 ((a) ![]() , almond; □, apricot;
, almond; □, apricot; ![]() , peach; ■, plum; (b)
, peach; ■, plum; (b) ![]() , Spain;
, Spain; ![]() , Greece; ■, Crete).
, Greece; ■, Crete).
Microsatellite data
We found one significant case of LD among loci (At006+At014, P=0.001; corrected α=0.0015). These two loci also showed significant deviations from HWE (both P<0.001; table 2) and 13 of the 15 un-amplifiable genotypes were also found at At006 and At014. Together, such patterns suggest the possibility of null alleles, so we removed these loci from the analyses presented here, though preliminary investigations of the complete data set suggest that their exclusion did not affect any of our conclusions.
At the remaining loci, as for the COI data, parasitoids from Spain and Greece show considerable geographic differentiation (‘Among Geographic Regions’: F CT=0.59, P<0.001; table 3a), but negative and insignificant ‘Among Host Plant’ group structure for the entire data set and separately within each region, as well as for analyses including or excluding apricot samples (table 3b–d). Pairwise F ST estimates (table 4) within regions were generally relatively low, ranging from 0.000 to 0.629, which only slightly overlapped with the high range of differentiation observed in pairs from different regions (0.553–0.798). In contrast, population pairs had very similar ranges of pairwise F ST, whether they were sampled from the same or from different host plant species (same: 0.000–0.798; different: 0.000–0.779). The majority of pairwise F ST estimates were significant (P<0.05); 30 of 31 insignificant estimates were for within-region population pairs (table 4). Poorly sampled populations did not exhibit F ST levels uncharacteristic of more well-sampled populations, though the degree of significance was somewhat reduced (table 4).
Table 3. AMOVAs of Aphidius transcaspicus microsatellite data grouped by (a) geographic region, (b) host plant/host plant with apricot samples removed, (c) host plant/host plant without apricot only within Spain and (d) host plant/host plant without apricot only within Greece. See Table 2 footnotes.

* See table 1 footnotes
Table 4. Pairwise estimates of genetic differentiation among Aphidius transcaspicus populations, measured by F ST (Weir & Cockerham, Reference Weir and Cockerham1984).

Sample sizes are given in parentheses next to population names, negative estimates are rounded to 0.000, estimates in bold are significant at P<0.05 and estimates in bold+italics are significant following strict Bonferroni correction (P<0.00026).
The extent of geographic differentiation is made more clear when pairwise F ST/(1−F ST) is plotted against geographic distance (fig. 3a). However, it is also clear that there is still a substantial degree of population structure within both Spain and Greece. The AMOVA showed significant differentiation ‘Among Populations within Regions’ for the geographically pooled samples (table 3a), and there was fairly high intra-regional differentiation when Spain (F ST=0.11, P<0.001) and Greece (F ST=0.22, P<0.001) were analyzed separately (table 3b, c). However, there was no significant correlation of pairwise genetic differentiation with distance within either region (fig. 3b, c), nor was there any indication from partial Mantel tests that populations sampled from different host plant species exhibited greater differentiation than populations from the same species at similar scales of spatial isolation. In fact, the positive r estimates suggest that populations from different plant species are actually more similar than those from the same plant species, significantly so in Greece (fig. 3c). While we suspect this significance is spurious (possibly due to the relatively large number of sampled almond populations), the analysis certainly provides no qualitative or quantitative support for greater genetic differentiation among populations on different host plants at any spatial scale.

Fig. 3. Correlations between F ST×(1−F ST)−1 calculated from the microsatellite data and geographic distance for populations of Aphidius transcaspicus. Each point represents a single pairwise comparison of two populations visualized for (a) the entire sample (•, both populations from Greece; ▿, populations from Greece and Spain; ■, both populations from Spain) and (b) within Spain and (c) within Greece. For (b) and (c), points are differentiated based on whether the population pair was sampled from (○) the same or (•) different plant species. Regression lines are drawn through points from (- - -) the same and (—) different plant species. Mantel tests for all populations showed no significant correlation of geographic and genetic distances in either region (Spain: r=−0.02, P=0.89; Greece: r=0.14, P=0.31). Partial Mantel tests using a host plant category variable (‘same Prunus sp.’=1, ‘different Prunus spp.’=0) suggested that populations from the same plant species were more genetically differentiated than from different plant species, significantly so in Greece (Spain: r distance=−0.02, P=0.71, r plant=0.24, P=0.16; Greece: r distance=0.14, P=0.60, r plant=0.33, P=0.01).
Both Bayesian and D C clustering analyses gave similar results. BAPS found that seven genetic clusters best explained the structure present in the microsatellite data (fig. 4). The clusters were cleanly split among the two geographic regions, with individuals from Greece assigned to {K1+K2+K3+K4} and those from Spain to {K5+K6+K7} (fig. 4). Once again, there was no obvious relationship between Prunus species and the inferred genetic structure within either region. In Greece (fig. 4a), for instance, K1 was found on almond, peach and plum; and K4 was found on all four host plants. Likewise, in Spain (fig. 4b), parasitoids belonging to K5 were present on all four host plant species. The remaining four clusters were present only in single populations (K2=Gb10almond, K3=Ga20almond, K6=S24apricot, K7=S28almond).

Fig. 4. Genetic structure of Aphidius transcaspicus inferred by the Bayesian admixture analysis implemented in BAPS, showing results for the most likely number of population partitions (K=7; log marginal likelihood=−1,878.87). Inferred population clusters were cleanly split into two groups by geographic region, with all 102 individuals from Greece assigned to (a) K1–K4 and all 88 individuals from Spain assigned to (b) K5–K7. Population clusters (a) within Greece and (b) within Spain are both organized by plant species (top) and by collection locality (bottom, see fig. 1) and divided into n vertical segments, where n is the number of individuals. The height of each colored segment corresponds to the posterior estimate of ancestry (admixture coefficient) to one or more Ks for that individual ((a) ■, K1;
![]() , K2;
, K2; ![]() , K3;
, K3; ![]() , K4; (b)
, K4; (b) ![]() , K5;
, K5;
![]() , K6; □, K7).
, K6; □, K7).
The NJ tree for inter-individual D C relationships also divided the samples into well-defined Spanish and Greek groups with good support (83% of bootstraps; fig. 5). In general, individuals from the different host plants were scattered throughout these two groups. There were a few possible ‘monophyletic’ host specific clusters apparent in the tree (marked by * in fig. 5). Closer inspection revealed that these were each comprised entirely of individuals from the same populations (e.g. S24apricot, S28almond; also identified as distinct by BAPS, fig. 4) rather than from multiple populations as would be expected for host associated differentiation.

Fig. 5. Unrooted neighbor-joining tree constructed among individual Aphidius transcaspicus microsatellites genotypes using the Cavalli-Sforza chord distance. Terminal branches are shaded according to the host plants from which parasitoids are reared, and the population composition of various groupings is provided (with the number of individuals). Support estimates are provided for branches that were observed in greater than 50% of bootstrapped distance matrices. Branches marked by a * represent groups of individuals that were also identified as distinct clusters by BAPS and discussed in the text (S24apricot, S28almond) (![]() , almond (thick black);
, almond (thick black); ![]() , apricot (thin gray);
, apricot (thin gray); ![]() , peach (thin black);
, peach (thin black); ![]() , plum (thick gray)).
, plum (thick gray)).
Discussion
Diversification of phytophagous insects in association with host plants is a remarkable and common evolutionary phenomenon (Funk et al., Reference Funk, Filchak and Feder2002), and the ways in which these patterns of differentiation may contribute to genetic structure at higher trophic levels is beginning to be appreciated (e.g. Stireman et al., Reference Stireman, Nason, Heard and Seehawer2006; Abrahamson & Blair, Reference Abrahamson, Blair and Tilmon2007). The unique pattern of parallel divergence between phytophagous insects and natural enemies in association with different host plants has been termed cascading (Stireman et al., Reference Stireman, Nason, Heard and Seehawer2006) or sequential (Abrahamson & Blair, Reference Abrahamson, Blair and Tilmon2007) host associated differentiation (henceforth, sequential host associated differentiation). Aphids in the genus Hyalopterus have been found to exhibit strong host associated differentiation on almond, peach and plum trees but with comparatively minor effects of geographic separation on overall genetic structure (Lozier et al., Reference Lozier, Roderick and Mills2007). In this study, we were interested in determining whether the specialized parasitoid A. transcaspicus showed similar patterns when reared from Hyalopterus species on these same set of host plants. If host associated differentiation was also driving diversification in A. transcaspicus, we would predict reduced gene flow among populations from different host plant species compared to populations from the same host plants. In the following, we first discuss how our results for A. transcaspicus are incompatible with this prediction and then explore possible factors that may limit the potential for sequential host associated differentiation in this parasitoid species.
Are there distinct host associated cryptic species within A. transcaspicus?
Our primary interest in this study was to test whether host plant associated speciation in Hyalopterus has triggered speciation in A. transcaspicus. For species-level host associated differentiation, individuals sampled from the same host plant species should, on average, share a more recent common ancestor than individuals from different host plant species, regardless of their geographic origins. In the absence of clear morphological differences or mating compatibility studies, one of the most useful ways to distinguish such a pattern is using Mallet's (Reference Mallet1995) genotypic clusters concept. Under this definition, groups of individuals are considered as species when they are analyzed with multiple genetic markers and consistently assigned to distinct genotypic clusters with no or few intermediates, thus strongly indicating the presence of reproductive isolation. Unlike for Hyalopterus, none of our molecular markers revealed a level of reproductive isolation consistent with host associated species for A. transcaspicus using this species concept.
First, the two most common COI haplotypes (A and B, 92% of samples) were detected on all four Prunus species, and all four haplotypes were separated only by a series of single base pair substitutions. This is in stark contrast to the six haplotypes found at the same COI region in Hyalopterus individuals collected from throughout the Mediterranean, which formed three distinct clades associated with almond, peach and plum and appear to have diverged several million years before present (4–8% divergence: Lozier et al., Reference Lozier, Roderick and Mills2007). Even if we exclude apricot samples to eliminate the potentially confounding influence of a shared Hyalopterus host plant, COI variation in A. transcaspicus does not approach the level of host plant association or degree of differentiation observed in its hosts. Second, AMOVA, Mantel tests, and both Bayesian and distance-based clustering analyses also failed to reveal any signature of host associated differentiation at the species level in the microsatellite data. Average genetic differentiation among parasitoids grouped by host plant was less than zero in the AMOVA (F CT=−0.11; table 3), and populations from different host plants were not more strongly differentiated than those from the same host plant at any spatial scale; if anything, the reverse was true (fig. 3). In comparison, grouping Hyalopterus populations by host plant explained much of the genetic variation present at microsatellite loci (F CT=0.2), while grouping by geographic region explained none of this variation (see Lozier et al., Reference Lozier, Roderick and Mills2007; table 3), a pattern that has been duplicated with morphological characters and has been deemed sufficient for the recognition of three distinct Hyalopterus species (Lozier et al., Reference Lozier, Foottit, Miller, Mills and Roderick2008). Our data show that similar cryptic host plant associated species are unlikely within A. transcaspicus in Spain and Greece.
Are there host races or biotypes within A. transcaspicus?
The question of whether A. transcaspicus exhibits intraspecific variation consistent with finer scale host associations is somewhat more difficult to address, in part because such intraspecific units are difficult to define and can encompass a large number of intermediate steps between panmictic populations and reproductively isolated species (Dres & Mallet, Reference Dres and Mallet2002). While we feel that our data are more consistent with a lack of any host-associated reproductive barriers in A. transcaspicus, we cannot completely rule out such a scenario.
It is always difficult to rule out type II error when failing to reject a null hypothesis. In this case, the small number of aphid mummies available from some localities, for example, may have limited our power to detect certain types of fine-scale population structure. Low power will be most problematic either for very recent population divergence or in the case of very low levels of genetic differentiation where populations still undergo considerable gene flow, as would be expected for host races (Dres & Mallet, Reference Dres and Mallet2002; Puebla et al., Reference Puebla, Bermingham and Guichard2008). Otherwise, individual genotypes from a poorly sampled population should still tend to group with those from more thoroughly sampled populations on the same host plant species because they will share more recent common ancestors. Given our clear rejection of species-level differences on different host plants, we can thus conclude that if host associated differentiation has occurred in A. transcpasicus it must be very recent – particularly given a probable 10–12 generations per year (unpublished data) – or at a very local scale that would have been difficult to observe with our sampling. While some host associated clustering of individuals was apparent (e.g. individuals from S24 on apricot, S28 on almond, Ga20 on almond, Gb10 on almond; fig. 4), these instances are equally consistent with other hypotheses given the restriction of these clusters to single localities. For example, geographic structure (e.g. for Ga20 sampled from the Peloponessus) or the sampling of closely related individuals within localities could both elevate interpopulation genetic variance. In general, individuals or populations from the same host plants were not more similar to each other than to individuals or populations from different host plants (figs 3–5), and in no cases did we find parasitoids from multiple populations of one host plant species clustering together to the exclusion of parasitoids from different host plants (figs 2, 4 and 5). Thus, there is little evidence to conclude that host associated differentiation is occurring within either Spain or Greece, indicating that neutral genetic differentiation at the within-region scale is also likely to be maintained by some process other than affinity of parasitoid genotypes to particular host plants.
Is there evidence for geographic differentiation within A. transcaspicus?
The hypothesis that seems best supported by our data is one where gene flow is limited largely by geographic separation of A. transcaspicus in Spain and Greece. Significant differentiation between the two regions was consistent for both molecular data sets but was most exceptional for the microsatellites (‘Among Region’ F CT=0.59), with some pairwise comparisons reaching F ST>0.75. This degree of structure is striking, indicating that parasitoids in these two regions have very different sets of alleles. This geographic differentiation could be confounded by the effect of sampling in different years. However, yearly fluctuation in allele frequencies seem unlikely to generate differentiation as large as that observed between Spain (2002) and Greece (2003, 2005) (tables 1 and 2). Furthermore, we detected no differences between the 2003 and 2005 Greek samples consistent with such temporal effects. It, thus, seems that gene flow between Spain and Greece is rare. Significant genetic structure within each country also indicates restricted dispersal at local scales, though the absence of increases in genetic differentiation with distance within regions further suggests that A. transcaspicus in Spain and Greece do not necessarily represent points within a single continuous population under IBD, but may be separated by more concrete barriers to dispersal. However, until intermediate populations are analyzed, we can make no definitive statements regarding range-wide models of population structure (Templeton, Reference Templeton1998).
In addition to the high degree of genetic differentiation, we observed an extremely low level of genetic polymorphism, particularly for the COI gene. This can be explained in part by reduced effective population sizes associated with haplodiploidy (Graur, Reference Graur1985), but a role for historical demographic scenarios cannot be excluded. For example, dispersal in insects associated with agriculture can be intimately tied to movements of crops by humans. Many species in the genus Prunus have been introduced to the Mediterranean only within the last 2000 years (Smartt & Simmonds, Reference Smartt and Simmonds1995), and it is entirely possible that both Hyalopterus and A. transcaspicus arrived concurrently. Recent non-equilibrium processes, such as founder events and subsequent range expansions, are consistent with the low level of polymorphism observed here, as well as with the failure to detect IBD within either Spain or Greece (Slatkin, Reference Slatkin1993). We are continuing to investigate A. transcaspicus populations sampled over a broader and more continuous scale in an attempt to better understand these intriguing patterns of geographic structure (Lozier et al., in prep.).
Sequential host associated differentiation and A. transcaspicus
Studies of phytophagous insects (Rhopalomyia solidaginis and Gnorimoschema gallaesolidaginis) and their natural enemies (Platygaster variabilis and Copidosoma gelechiae) on goldenrod (Solidago spp.) currently provide the best empirical evidence for sequential host associated differentiation in nature (Nason et al., Reference Nason, Heard and Williams2002; Stireman et al., Reference Stireman, Nason, Heard and Seehawer2006). Studies of Cotesia parasitoids of checkerspot butterflies also lend some support for sequential host associated differentiation, though the importance of the host plant in this radiation is not entirely clear (Kankare et al., Reference Kankare, van Nouhuys and Hanski2005). However, there are still few examples of sequential host-associated differentiation in the literature, and it appears that this mechanism will not apply to all systems (Stireman et al., Reference Stireman, Nason, Heard and Seehawer2006; Abrahamson & Blair, Reference Abrahamson, Blair and Tilmon2007). For example, in another study on Solidago, Cronin & Abrahamson (Reference Cronin and Abrahamson2001) were unable to detect diversification of the parasitoid Eurytoma gigantea in response to host race formation in Eurosta gall flies. Similarly, a study of parasitoids of green cloverworm on alfalfa and soybean failed to detect reproductive isolation using AFLPs (Medina, Reference Medina2005). We are unaware of any previous studies testing specifically for sequential host associated differentiation in the Aphidiinae. While some aphidiines certainly consist of cryptic host-specific complexes (Tremblay & Pennacchio, Reference Tremblay, Pennacchio and Gupta1988; Atanassova et al., Reference Atanassova, Brookes, Loxdale and Powell1998), studies of host specialization have typically examined parasitoids that attack relatively unrelated aphids rather than recently evolved cryptic species. More work is needed to test the generality of sequential host associated differentiation, and host associated differentiation overall, so that specific factors that either promote or inhibit diversification at different trophic levels and in different taxonomic groups can be identified (Abrahamson & Blair, Reference Abrahamson, Blair and Tilmon2007).
Given that aphidiines possess characteristics that could promote host associated differentiation (see Introduction; Abrahamson & Blair, Reference Abrahamson, Blair and Tilmon2007), what factors, apart from statistical concerns, might explain the apparent failure of A. transcaspicus to diverge with Hyalopterus? An important attribute of parasitic organisms is that their population structure is governed both by intrinsic properties such as dispersal and by extrinsic dynamics of host populations (Huyse et al., Reference Huyse, Pulin and Theron2005). Aphid parasitoids, for example, can disperse as adults, but also as eggs or larvae within parasitized migratory hosts (Feng et al., Reference Feng, Chen, Shang, Ying, Shen and Chen2007). Each summer, Hyalopterus populations obligately disperse from their primary Prunus hosts to the secondary host plant (e.g. Phragmites), followed in autumn by a return to primary hosts for sexual reproduction and overwintering. While the three Hyalopterus species are largely specific to their primary host plants (Lozier et al., Reference Lozier, Roderick and Mills2007), they co-occur on Phragmites (unpublished data). For A. transcaspicus, selection for responses to primary host plant cues (e.g. for mate or host location: Storeck et al., Reference Storeck, Poppy, van Emden and Powell2000) would be ineffective at maintaining reproductive isolation when parasitoids emerge on this shared host plant. Similar patterns could arise when parasitized aphids settle on host plants shared by all three Hyalopterus species, such as apricot, or make the occasional ‘wrong’ host plant choice (Lozier et al., Reference Lozier, Roderick and Mills2007). Theoretical investigations of the conditions facilitating stable divergence of a predator following ecological divergence in the prey suggest that this secondary divergence will only occur if the new prey species are sufficiently ecologically distinct and if assortative mating is sufficiently strong (Dieckmann & Doebeli, Reference Diekmann, Doebeli, Dieckmann, Doebeli, Metz and Tautz2004). Otherwise, the predator remains a generalist. The phenomena discussed above could promote gene flow among parasitoid populations whose ancestors had emerged on different host plants several generations earlier; and, in the face of such gene flow, ecological differences among Hyalopterus or Prunus species (e.g. quality of chemical cues, physiological response to parasitoid attack) may simply provide insufficient disruptive selection to allow the evolution of specialization and reproductive isolation below the level of host genus (Hufbauer, Reference Hufbauer2001; Bush & Butlin, Reference Bush, Butlin, Dieckmann, Doebeli, Metz and Tautz2004).
Another possibility is that the opportunity for sequential host associated differentiation in A. transcaspicus may be affected by resource limitation associated with fluctuations in the availability of the different Hyalopterus species. The evolution of specialization can benefit parasitoids by increasing efficiency and virulence or by reducing competition, but could come at the cost of reduced plasticity when the preferred host is rare or unavailable (Antolin et al., Reference Antolin, Bjorksten and Vaughn2006). Field trips to Mediterranean localities in multiple years have revealed marked differences in aphid abundance on the different Prunus species from year to year (N. Mills, personal observation). If encounters with the different Hyalopterus species are sufficiently unpredictable due to this variation, the maintenance of a partially generalist lifestyle could be favored (Berlocher & Feder, Reference Berlocher and Feder2002; Lapchin, Reference Lapchin2002; Antolin et al., Reference Antolin, Bjorksten and Vaughn2006).
In summary, mtDNA and microsatellite data and several analytical approaches indicate a primary role for spatial factors, rather than host plant associations, in maintaining genetic structure in A. transcaspicus and that, at the species level, A. transcaspicus should be considered a generalist with respect to its Hyalopterus hosts. With regard to biological control of H. pruni on dried plum in North America, our results suggest that targeting A. transcaspicus populations from aphids on P. domestica may be less important than considering differences among regional populations. For example, given the broad geographic distribution and high degree of genetic structure in its native range, regional A. transcaspicus populations are likely to possess unique adaptations to different environmental pressures. Selecting parasitoids from areas with climates most similar to California's Central Valley may, thus, be crucial for establishment, as has been the case for past biological control programs (Messenger & van den Bosch, Reference Messenger, van den Bosch and Huffaker1971; van den Bosch et al., Reference van den Bosch, Hom, Matteson, Frazer, Messenger and Davis1979).
That said, it should be stressed that an absence of host plant or host associated population structure at neutral genetic markers does not necessarily indicate lack of differential fitness or behavioral variation in host preference in local populations. Little gene flow is needed to overcome the effects of genetic drift at neutral loci while selected loci can remain structured (Slatkin, Reference Slatkin1987; Feder et al., Reference Feder, Berlocher, Opp, Mopper and Strauss1998). Indeed, host and host plant preferences have been demonstrated in several aphid parasitoids that showed no host associated differentiation at neutral genetic markers (Daza-Bustamante et al., Reference Daza-Bustamante, Fuentes-Contreras, Rodriguez, Figueroa and Niemeyer2002; Baer et al., Reference Baer, Tripp, Bjorksten and Antolin2004; Antolin et al., Reference Antolin, Bjorksten and Vaughn2006); and the far greater abundance of A. transcaspicus on almond trees suggests that aphids from this host plant may be preferred, at least in the regions sampled for this analysis. We suggest that future research effort to address the question of host preference in this parasitoid may be better spent on behavioral and ecological experiments rather than on attempts to identify the low levels of population structure that would likely be present among potential host races. If host preferences can be established experimentally, then further tests for fine-scale reproductive isolation would be warranted. Further analyses of geographic population structure throughout the Mediterranean, however, may help determine factors that can explain the extraordinarily high genetic differentiation and low genetic diversity that we have observed for Spain and Greece. Together, such studies may provide additional insights into both the evolution of host use and diversification in this parasitoid and may be of great practical importance for the selection employment of the most effective A. transcaspicus populations for management of Hyalopterus where it is a pest.
Acknowledgements
We gratefully recognize funding support from the EPA STAR program, the UC IPM Exotic Pest and Disease Research Program, USDA NRI and the University of California Agriculture Experiment Station. We also thank Dr Petr Starý, Sean Schoville and anonymous reviewers for helpful comments and discussion.











