Introduction
The genus Epitrix Foudras (Coleoptera: Chrysomelidae: Alticinae) comprises nearly 180 species worldwide. Most of the species occur in the neotropics (130) and only 12 and 17 species are known from North America and Europe, respectively (Doeberl, Reference Doeberl2000).
Epitrix species feed mainly on plants from the family Solanaceae (Doguet, Reference Doguet1994), though they may feed on other plant families when their preferential host is not available (e.g., Chenopodiaceae, Cucurbitaceae, Fabaceae (European and Mediterranean Plant Protection Organization (OEPP/EPPO), 2005, Table 1)). Five North American native species have been reported to feed on potatoes (Solanum tuberosum L.): Epitrix cucumeris (Harris), Epitrix tuberis Gentner, Epitrix similaris Gentner, Epitrix subcrinita (LeConte) and Epitrix hirtipennis (Melsheimer), and the two first are considered serious pests of potato crops (Gentner, Reference Gentner1944; Hoy et al., Reference Hoy, Boiteau, Alyokhin, Dively, Alvarez and Johnson2007, Table 1).
Table 1. Summary of available data on the five Epitrix species reported to feed on potatoes (S. tuberosum) in North America and Europe.

Epitrix species being morphologically very similar, they are often considered a pest complex. Therefore, data on species distribution, host range and damage should be taken with caution.
* Detailed distributions can be found in OEPP/EPPO (2010b).
Adults feed on foliage, producing numerous small round holes (1.0–1.5 mm diameter, ‘shot-hole’ pattern) (OEPP/EPPO, 2011). Young plants and seedlings are particularly susceptible to attacks and heavy infestations may cause plant stunting. However, damage caused by adults is rarely economically important. When present in large numbers, soil dwelling Epitrix larvae, which feed on roots, stolons and tubers, may be responsible for more serious damage and important economic losses (Gentner, Reference Gentner1944; Morrison et al., Reference Morrison, Gentner, Koontz and Every1967). For example, E. tuberis larvae burrow into tubers, leaving roughened trails on the surface, or tiny tunnels extending as far as 1.5 cm into the tuber flesh (‘worm track’ damage) that are still evident after peeling. Tunnels may cause deep cracks, rough and pimply skin and sometimes distortion of the tuber. One or two larvae can do enough damage to make a tuber unmarketable (OEPP/EPPO, 1989). Furthermore, larvae may act as vectors of bacteria (e.g., Streptomyces spp.), viruses (e.g., Andean Potato latent tymovirus, APLV) and fungal pathogens (Verticillium dahliae, Fusarium coeruleum and Thanatephorus cucumeris), which may increase financial losses result from damaged crops (OEPP/EPPO, 2005, 2010a; Vreugdenhil et al., Reference Vreugdenhil, Bradshaw, Gebhardt, Govers, MacKerron, Taylor and Ross2007, Table 1).
Epitrix species are morphologically highly similar, which makes them difficult to identify for non-specialists. Adults are tiny pubescent beetles, generally ranging in size from 1.5 to 2 mm. Identification of the species is mostly based on the observation of the genitalia of both sexes and requires a high level of expertise. There is no comprehensive recent revision of the genus and consequently only local and partial keys are available (Seeno & Andrews, Reference Seeno and Andrews1972; Doeberl, Reference Doeberl2000; Warchalowski, Reference Warchalowski2003). No diagnostic characters have been reported for the immature life stages. Difficulties in distinguishing between Epitrix species limit our knowledge of their biology and render their management particularly challenging (Boavida & Germain, Reference Boavida and Germain2009). Epitrix species are often considered a pest complex (OEPP/EPPO, 2010b). In many records, specimens are not identified to species, which makes available data on species distribution and host range not fully reliable. Field identification is generally based on type of damage to tubers or foliage. However, as a systematic survey on Epitrix species in North America has not been attempted since Gentner (Reference Gentner1944), confusion of symptoms because of species misidentification may exist and pest status of some species may be re-assessed in the future (Boavida & Germain, Reference Boavida and Germain2009).
E. subcrinita and E. tuberis, which have not been detected in Europe so far, are listed as quarantine pests (A1 list) by OEPP/EPPO (1989). Following the recent report of E. cucumeris and E. similaris in Portugal (Boavida & Germain, Reference Boavida and Germain2009), those two species have been listed as quarantine pests (A2 list) by EPPO. Pest risk analysis has shown that E. cucumeris and E. similaris could spread and find environmental factors suitable for establishment in most European countries (OEPP/EPPO, 2010b). Finally, E. hirtipennis, which was introduced to Europe in 1984 (Italy), is not considered a serious pest of potato crops in the European countries where it is established (Boavida & Germain, Reference Boavida and Germain2009).
With 62 millions tons produced per year (2008 data), potato production in the European Union (EU, 27 member states) is ranked second in the world after China (Schwartzmann, Reference Schwartzmann and Schepers2010). Specifically, in the EU-5 zone comprising the United Kingdom, the Netherlands, Belgium, Germany and Northern France, the production and international trade of ware potatoes (usually as washed commodities) generate a sales value of about $4.3 billion a year (Schwartzmann, Reference Schwartzmann and Schepers2010).
By reducing the quality and marketable yield of ware potatoes and requiring additional insecticide applications for pest control, tuber-damaging flea beetles represent a considerable threat for the European agricultural economy and environment. For example, once Epitrix species have established, economic losses for English main crop potato growers could be as high as $15–60 millions per year (Fera, 2012). Those estimates include increased costs of additional insecticide applications and revenue losses because of reduction in quality and marketable yield. However, loss of export markets for ware and seed potatoes are not included, hence losses may even be underestimated. Human-mediated spread seems more likely than natural spread, but flight distances are poorly known (OEPP/EPPO, 2010b). The most likely pathways for introduction and spread are through soil attached to roots or tubers where larvae, pupae or overwintering adults may occur (Cusson et al., Reference Cusson, Vernon and Roitberg1990; OEPP/EPPO, 2010b). Currently, the EU Standing Committee on Plant health is working altogether with member state plant protection services on emergency measures to prevent the introduction of E. subcrinita and E. tuberis and the spread of E. cucumeris and E. similaris in Europe (J.-F. Germain, personal communication). Therefore, a tool to reliably identify all developmental stages of potato flea beetles occurring in Europe and North America is urgently needed.
Here, we compared the effectiveness of the mitochondrial cytochrome c oxidase I (COI) standard barcode fragment (Hebert et al., Reference Hebert, Cywinska, Shelley and deWaard2003) and the nuclear internal transcribed spacer 2 (ITS2) in differentiating among six morphospecies of Epitrix occurring in Europe and North America. Indeed, mitochondrial introgression, which can mislead species identification (see e.g., Frezal & Leblois, Reference Frezal and Leblois2008) has been reported in several groups of Chrysomelidae (e.g., Gomez-Zurita & Vogler, Reference Gomez-Zurita and Vogler2006; Campbell et al., Reference Campbell, Clark, Clark, Meinke and Foster2011). As no mitochondrial introgression was detected, we then used COI sequences and tested the potential of the polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) approach to rapidly distinguish among Epitrix species.
Materials and methods
Taxonomic sampling and morphological identification
We sampled 126 specimens of Epitrix from several localities across Canada (British Columbia and Prince Edward Island), Costa Rica, Portugal (including the Azores, which lies some 1500 km west of the coast of Portugal) and the United States (California, Kansas and Wisconsin) between April 2010 and August 2011. Samples were mostly collected on S. tuberosum L., though several specimens were collected on Physalis longifolia Nutt. (Long-leaf groundcherry) and Capsicum sp. (table 2). Specimens were collected alive and stored in 95% ethanol. Specimens were mostly collected as adults, though we also assessed the reliability of our molecular tools by testing assignment to species of a few larvae (table 2). Adult specimens were identified to species by J.-F.G. based on examination of habitus and genitalia, following Seeno & Andrews (Reference Seeno and Andrews1972), Doeberl (Reference Doeberl2000), Warchalowski (Reference Warchalowski2003) and OEPP/EPPO (2011). Preparations of genitalia were made using the following protocol: genitalia were dissected from specimens using two needles, gently heated in a 10% KOH solution for 20 min, transferred to water (20 min), dehydrated using 70% EtOH and 100% EtOH (5 min each), transferred to lavender oil (for at least 10 min) and slide-mounted in a drop of Canada balsam for permanent storage. Slides were deposited at ANSES-LSV, Montferrier-sur-Lez, France.
Table 2. List of Epitrix specimens included in this study.
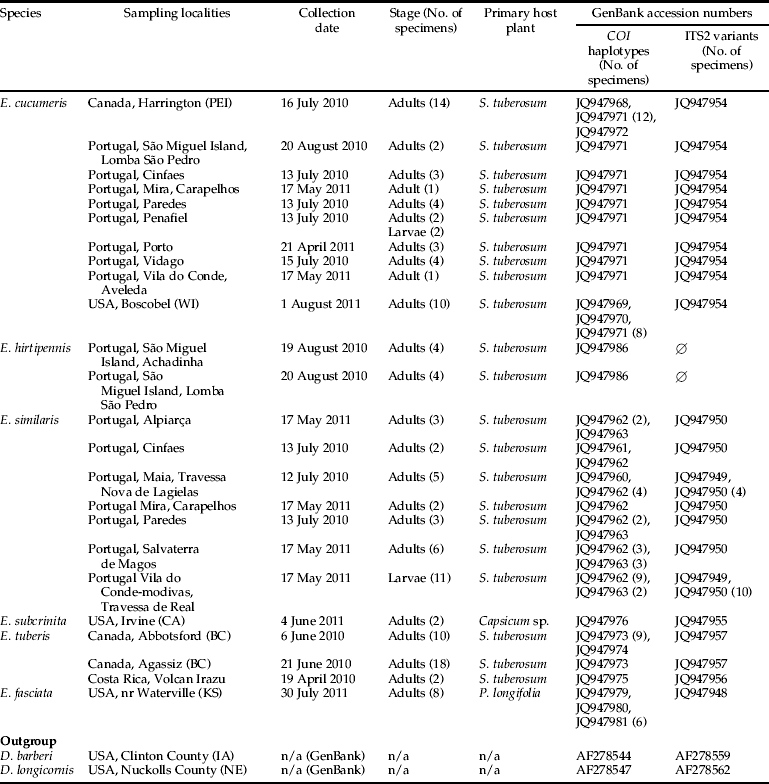
Abbreviations used: BC, British Columbia; CA, California; IA, Iowa; KS, Kansas; NE, Nebraska; PEI, Prince Edward Island; WI, Wisconsin.
DNA extraction, amplification and sequencing
Genomic DNA was isolated using the Qiagen DNeasy kit (Hilden, Germany) following the manufacturer's protocol without destruction of the specimens, to allow subsequent examination of morphology.
This study is part of an ongoing project (Quarantine Barcode of Life, QBOL), which aims at developing diagnostic resources to enable molecular identification of quarantine arthropod pests for Europe (Bonants et al., Reference Bonants, Groenewald, Rasplus, Maes, de Vos, Frey, Boonham, Nicolaisen, Bertacini, Robert, Barker, Kox, Ravnikar, Tomankova, Caffier, Li, Armstrong, Freitas-Astua, Stefani, Cubero and Mostert2010). About 300 species from seven different arthropod orders are sequenced not only on the standard 658 bp region of the COI gene (Hebert et al., Reference Hebert, Cywinska, Shelley and deWaard2003) but also on ITS2. The choice of ITS2 was motivated by the fact that primers could be designed in regions that were conserved across many taxa (i.e., 5.8S and 28S rRNA) and that ITS2 often exhibits more intra-specific variability than other nuclear markers.
To achieve our general goal, efforts were taken to amplify all species following a single protocol. Primers were designed as follows: for both COI and ITS2, we aligned as many COI and ITS2 sequences as possible from arthropod species, found on GenBank. For COI, we started from the classical Folmer et al. (Reference Folmer, Black, Hoeh, Lutz and Vrijenhoek1994) primers and designed more optimal primers by degenerating appropriate sites to increase amplification success. ITS2 primers were designed in regions that were conserved across all taxa, with appropriate degenerate sites to increase amplification success. Furthermore, instead of using pairs of conventional degenerate primers (one forward and one reverse), COI and ITS2 were amplified using primer cocktails, to further increase amplification success (Ivanova et al., Reference Ivanova, Zemlak, Hanner and Hebert2007). Finally, as PCR products were amplified from primer cocktails, primers were M13-tailed to allow sequencing (table 3, tails being highlighted in pale grey). Indeed, M13-tailed primers have proven more effective, allowing longer sequencing reads with more overlap (Ivanova et al., Reference Ivanova, Zemlak, Hanner and Hebert2007).
Table 3. PCR primer cocktails used in this study.
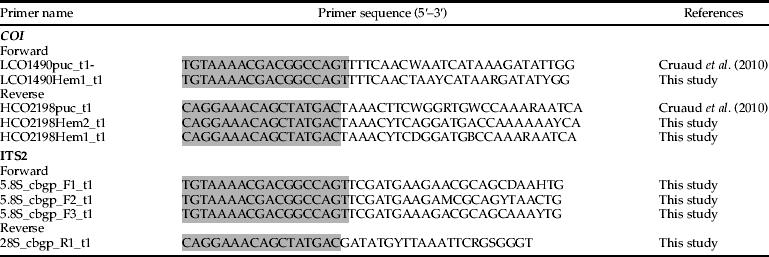
M13 tails from Ivanova et al. (Reference Ivanova, Zemlak, Hanner and Hebert2007) are highlighted. Original references for the untailed versions of each primer are provided.
All PCRs were performed using the following reagents in a 25 μl reaction volume: 4 μl of genomic DNA (25–200 ng), 15.875 μl of ultra pure water, 2.5 μl of 10 × PCR buffer (final concentration = 1×), 0.5 μl of 25 mM MgCl2 (0.5 mM), 0.5 μl of each 10 μM primer cocktail (0.2 μM), 1 μl of each 2.5 mM dNTP (0.1 mM), 0.125 μl of 5 units Taq DNA Polymerase (Qiagen, Hilden, Germany) (0.625 unit). PCR conditions for COI were: 94 °C for 2 min, five cycles of 94 °C for 30 s, 45 °C for 40 s and 72 °C for 60 s, followed by 35 cycles of 94 °C for 30 s, 51 °C for 40 s and 72 °C for 60 s, with a final extension at 72 °C for 10 min. PCR conditions for ITS2 were: 94 °C for 2 min, five cycles of 94 °C for 30 s, 45 °C for 60 s and 72 °C for 90 s, followed by 35 cycles of 94 °C for 30 s, 55 °C for 60 s and 72 °C for 90 s, with a final extension at 72 °C for 10 min. PCR products were visualized on a 2% agarose gel using an E-Gel96 Pre-cast Agarose Electrophoresis System (Invitrogen, Paisley, UK). Unpurified PCR products were sent to Eurofins MWG Operon (Ebersberg, Germany) for sequencing using M13F (−21) 5′-TGTAAAACGACGGCCAGT-3′) and M13R (−27) 5′-CAGGAAACAGCTATGAC-3′ primers (Ivanova et al., Reference Ivanova, Zemlak, Hanner and Hebert2007), which correspond to the ‘tails’ added to the PCR primers.
Both strands for each overlapping fragment were assembled using CodonCode Aligner v 3.7.1.1 (CodonCode Corporation, Dedham, Massachusetts, USA). Divergent haplotypes obtained for each marker were deposited in GenBank (table 2).
Sequence data analyses
Sequence alignment
All gene regions were aligned with MAFFT 6.864 (Katoh et al., Reference Katoh, Kuma, Toh and Miyata2005) using the L-INS-i option. COI alignment was translated to amino acids using MEGA 4 (Tamura et al., Reference Tamura, Dudley, Nei and Kumar2007) to detect frame-shift mutations and premature stop codons, which may indicate the presence of pseudogenes.
Distance analyses
Pairwise nucleotide sequence divergences were calculated using a Kimura 2-parameter model of substitution (Kimura, Reference Kimura1980) in MEGA 4, using the ‘pairwise-deletion’ of gaps option.
Phylogenetic reconstruction
The most appropriate model of evolution for each gene region was identified using the Akaike information criterion implemented in MrAIC.pl 1.4.3 (Nylander, Reference Nylander2004). We performed maximum likelihood (ML) analyses of the two gene regions using MPI-parallelized RAxML 7.2.8 (Stamatakis, Reference Stamatakis2006a). GTRCAT approximation of models was used for ML bootstrapping (Stamatakis, Reference Stamatakis2006b) (1000 replicates). Analyses were conducted on a 150 cores Linux Cluster at CBGP. COI and ITS2 sequences from Diabrotica barberi Smith & Lawrence, 1967 and Diabrotica longicornis (Say, 1824) were downloaded from GenBank and used as outgroups (table 2).
PCR–RFLP analyses
Owing to failure to amplify orthologous ITS2 sequences for E. hirtipennis and given that no mitochondrial introgression was detected between Epitrix species, PCR–RFLP analyses were carried out on COI only. Restriction patterns were predicted using BioEdit (Hall, Reference Hall1999) and TaqI was selected to allow discrimination of the six species. COI amplicons from all tested species and haplotypes were subjected to TaqI restriction activity at 65 °C for 3 h. Ten microlitres of PCR product were digested with 5.0 units of TaqI (Promega, Madison, Wisconsin, USA), 1 × buffer E and sterile distilled H2O in a 15 μl reaction volume. The restriction fragments were separated by electrophoresis on 1.5% agarose gel at 110 V for 90 min and stained with ethidium bromide.
Results
Taxonomic sampling and morphological identification
Adult specimens were sorted into six morphospecies (table 2). Specimens collected in Portugal (49) were identified either as E. cucumeris (41%), E. similaris (43%) or E. hirtipennis (16%, collected only in the Azores).
Amplification success and sequence data
COI (658 bp) and ITS2 (691 aligned bp) were successfully amplified from all adult specimens and larvae. Alignment of COI was straightforward owing to a lack of length variation and no stop codons or frame shifts were detected. Alignment of ITS2 revealed that a divergent paralog was sequenced from all specimens of E. hirtipennis. A second attempt to amplify orthologous sequences led to the same result and ITS2 sequences from E. hirtipennis were consequently excluded from the analysis.
Distance and phylogenetic analyses
With 17 haplotypes identified, COI was more variable than ITS2, for which only seven variants were detected (table 2). K2P pairwise distances between Epitrix specimens (as per cent sequence divergence) are summarized in table 4. The intra-specific K2P distance range for COI was 0.00–1.39% (mean 0.15%), while the inter-specific distances ranged from 7.29% to 24.09% (mean 17.55%). The intra-specific K2P distance range for ITS2 was 0.00–0.18% (mean 0.01%) while the inter-specific distances ranged from 0.36% to 24.86% (mean 8.23%). Whatever the marker used, the minimum inter-specific divergence exceeded the maximum intra-specific divergence for all species.
Table 4. Kimura two-parameter pairwise distances (percentage) between specimens of Epitrix species.
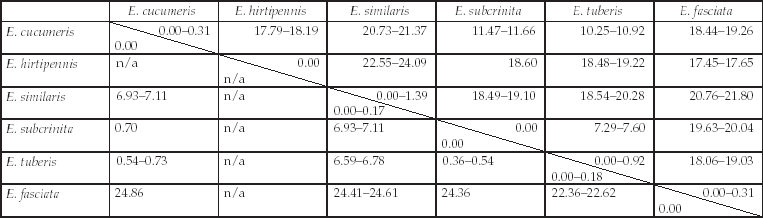
COI distances are reported in the upper triangular matrix and ITS2 distances (calculated using the ‘pairwise deletion’ of gaps option) are reported in the lower triangular matrix. Off-diagonal entries: ranges of pairwise distances between samples among Epitrix species (inter-specific divergence); on diagonal entries: ranges of pairwise distances within Epitrix species (intra-specific divergence). n/a: non applicable.
Models chosen by MrAIC were as follows: GTR + I+Γ for COI and GTR + Γ for ITS2. Given that α and the proportion of invariable sites cannot be optimized independently from each other (Gu, Reference Gu1995) and following Stamatakis’ personal recommendations (RA × ML manual, 2006a), we used GTR + Γ with four discrete rate categories for both COI and ITS2. Phylogenetic analyses of COI and ITS2 (fig. 1) recovered the same well-supported clusters of sequences, which corresponded to morphologically delineated species. E. tuberis was the sole species to show two geographical clusters of haplotypes (Canada versus Costa Rica).
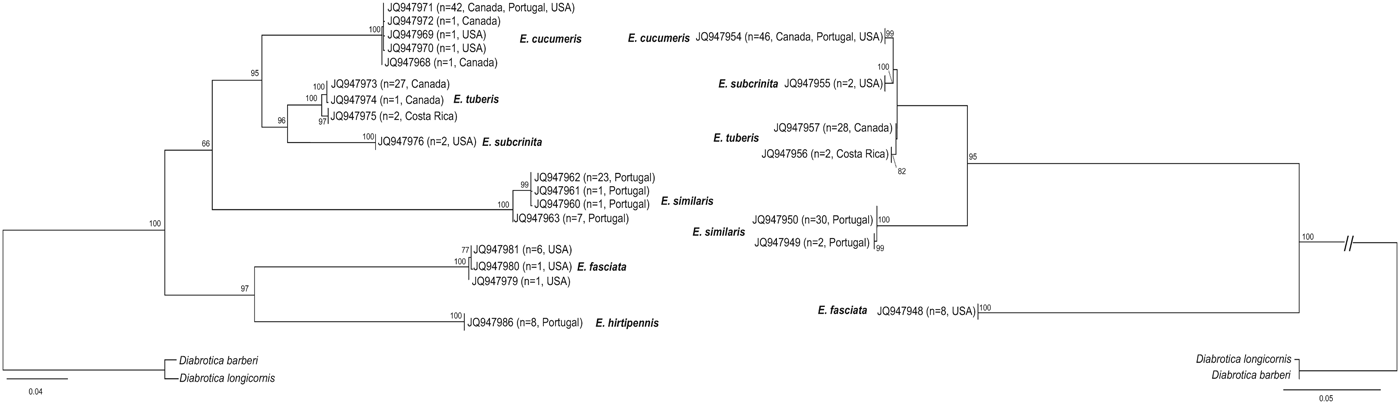
Fig. 1. Ml trees from the analyses of COI (left) and ITS2 (right) sequences. Specimens with identical haplotypes are collapsed to a single terminal node (GenBank accession numbers, number of specimens and geographical origin are indicated for each haplotype; details are provided in table 2). Bootstrap supports higher than 60% are indicated at nodes.
PCR–RFLP analyses
RFLP profiles for the 658 bp fragments of COI digested with TaqI are shown in fig. 2. In all species, alternate haplotypes produced unique RFLP patterns. TaqI activity produced four restriction fragments for E. hirtipennis (338, 260, 251 and 151 bp), three restriction fragments for Epitrix fasciata (609, 154 and 151 bp), and two restriction fragments for E. similaris (505 and 325 bp), E. subcrinita (573 and 257 bp) and E. tuberis (676 and 154 bp). No TaqI cleavage was detected for E. cucumeris. The two smallest fragments for E. fasciata (154 and 151 bp) were hardly distinguishable on the gel, though without precluding species identification.

Fig. 2. RFLP profiles for a 658 bp fragment of COI digested with the restriction enzyme TaqI in the six species of Epitrix. GenBank IDs of the different haplotypes are indicated on lanes (see table 2 for details). Far right lane contains GeneRuler™ DNA ladder Mix (Fermentas, Walthman, Massachusetts, USA), with the first 11 fragments ranging in size from 100 bp to 1000 bp in 100 bp increments.
Discussion
Since reference libraries validated by taxonomists are available, COI DNA barcodes (Hebert et al., Reference Hebert, Cywinska, Shelley and deWaard2003) have been proposed as efficient diagnostic tools for biosecurity (e.g., Armstrong & Ball, Reference Armstrong and Ball2005; de Waard et al., Reference de Waard, Mitchell, Keena, Gopurenko, Boykin, Armstrong, Pogue, Lima, Floyd, Hanner and Humble2010; Floyd et al., Reference Floyd, Lima, deWaard, Humble and Hanner2010). Successful barcode identification requires intra-specific variability being markedly lower than inter-specific variability (Hebert et al., Reference Hebert, Stoeckle, Zemlak and Francis2004). When mitochondrial introgression occurs, which has been reported in several groups of Chrysomelidae (e.g., Gomez-Zurita & Vogler, Reference Gomez-Zurita and Vogler2006; Campbell et al., Reference Campbell, Clark, Clark, Meinke and Foster2011), this assumption is broken. Intra- and inter-specific variation can also overlap when species are not reciprocally monophyletic, for example when there is incomplete lineage sorting owing to the retention of ancestral polymorphism (Funk & Omland, Reference Funk and Omland2003).
Here, we show that COI DNA barcodes should be valuable in routine identifications of all developmental stages of potato flea beetles occurring in Europe and North America. Morphospecies of Epitrix were indeed well differentiated by COI DNA barcodes. Minimum inter-specific divergence largely exceeded maximum intra-specific divergence and all species were recovered as reciprocally monophyletic by our phylogenetic analyses. Furthermore, comparison with ITS2 genetic clusters revealed no mitochondrial introgression between species. It is noteworthy that, while widely distributed in North America and established in the Azores, Italy, Greece, Bulgaria and Turkey, E. hirtipennis was sampled from the Azores only. One could therefore expect slight differences between sequences from our samples and those from other geographic regions, but this should not preclude species identification, though this needs to be formally established. Indeed, intra-specific distances were overall very low. For example, for E. cucumeris, which was sampled in Portugal (including the Azores), Canada and the United States intra-specific distances did not exceed 0.32% for both COI and ITS2.
While amplification and sequencing of COI were straightforward for both adults and larvae in every species, we failed to amplify orthologous ITS2 sequences for E. hirtipennis. This indicates that COI should be preferred over ITS2 when developing a reference barcode database for identification and monitoring of Epitrix species. Providing a molecular diagnostic tool for the identification of potato flea beetles occurring in other parts of the world or other Epitrix species was not the purpose of this study. However, primer cocktails and PCR conditions provided here should help in developing such tools. As mentioned in the Materials and methods section, this study is part of the QBOL project, which aims at developing diagnostic resources to enable molecular identification of quarantine arthropod pests for Europe (Bonants et al., Reference Bonants, Groenewald, Rasplus, Maes, de Vos, Frey, Boonham, Nicolaisen, Bertacini, Robert, Barker, Kox, Ravnikar, Tomankova, Caffier, Li, Armstrong, Freitas-Astua, Stefani, Cubero and Mostert2010). Information regarding barcoded specimens (including those used in the present study) as well as barcode sequences themselves are deposited in an Internet-based database system, Q-bank (http://www.q-bank.eu/arthropods/). A BLAST tool allows online identification of unknown specimens by querying the sequence database.
To best match the available lab equipment and management objectives, we also developed an RFLP-based diagnostic method and showed that unambiguous species discrimination can be achieved by using the sole restriction enzyme TaqI on COI PCR products. Thanks to significant reduction in processing time and cost, this RFLP-based identification tool may allow for more extensive sampling strategies and hence more efficient field monitoring surveys.
Prior to this study, identification of Epitrix species was based on examination by expert taxonomists of morphological characters of adult specimens, especially genitalia. No diagnostic characters were available for larvae and non-specialists relied exclusively on plant damage to identify species. However, plant damage-based diagnosis can be misleading as species misidentification might have resulted in confusion of symptoms. For example, Boavida & Germain (Reference Boavida and Germain2009), recently raised the possibility that E. tuberis and E. similaris, which are sympatric in California (Seeno & Andrews, Reference Seeno and Andrews1972) and resemble each other in their external morphology (Gentner, Reference Gentner1944), may have been confused. Indeed, in Portugal, where it has been recently introduced, E. similaris seems responsible for ‘worm track damage’ to tubers usually attributed to E. tuberis (R. Oliveira, personal communication). By enabling the identification of larvae, molecular diagnostic tools make possible linking Epitrix species to observed damage, which should improve our knowledge of species biology. Furthermore, the tool provided here should clarify Epitrix distribution and host range, two capacities that are also particularly important in the management of these pest species in Europe and North America. Our study confirms that E. tuberis and E. subcrinita have probably not been introduced to Europe yet (Boavida & Germain, Reference Boavida and Germain2009). Indeed, including this study some hundreds of Epitrix specimens have already been sampled in Europe, but not one specimen of these two species has been detected so far. Besides potato flea beetles, we also sampled E. fasciata, the Southern tobacco flea beetle, which preferentially develop on Nicotiana tabacum L. E. fasciata has been recently introduced to the Azores but is not considered a pest of potato (Boavida & Germain, Reference Boavida and Germain2009). However, E. fasciata and E. hirtipennis being morphologically highly similar, molecular barcodes should facilitate distinguishing these two species.
During the final stages of review of this manuscript, the European commission published a decision (212/270/EU) to prevent the introduction of E. tuberis and E. subcrinita in Europe and limit the spread of E. cucumeris and E. similaris (Official Journal of the European Union, 2012). By enabling the identification of Epitrix specimens at different periods of the life cycle, this diagnostics tool should help member states conducting survey for the presence of potato flea beetles on potato crops as well as other host plant species, as recommended by the European commission. Fast and accurate detection of Epitrix potato flea beetles would help study their potential spread and contribute to their management with minimal disruption to Solanaceae trade.
Acknowledgments
We thank L. Lesage (Agriculture and Agri-Food Canada, Ottawa, Ontario), B. Vernon (Agriculture and Agri-Food Canada, Agassiz, British Columbia), R. Oliveira (Germicopa-Portugal) and P.E. Hanson (Universidad de Costa Rica) for contributing samples. We also thank S. Doguet (France) and M. Döberl (Germany) for cross-checking identification of E. cucumeris and E. similaris in Europe. We also thank Charles-Antoine Dedryver (INRA, France) for his careful reading of this manuscript and thoughtful suggestions for revisions. This work was based on financial supports received from the QBOL project allocated by the European Union (7th FW, 2009–2012) and the BIOFIS project (number 1001-001) allocated by the French Agropolis Fondation (RTRA – Montpellier) to J.Y.R.








