Introduction
It is well established that vitellogenesis is associated with significant accumulation of yolk proteins. These are usually phospholipoglycoproteins and play a key role in embryogenesis by providing nutrient sources such as lipids, free amino acid, carbohydrates, carotenoids, and minerals (Adams et al., Reference Adams, Filipi and Yi2002; Khalaila et al., Reference Khalaila, Peter-Katalinic, Tsang, Radcliffe, Aflalo, Harvey, Dwek, Rudd and Sagi2004) to the growing oocytes, embryos (Lee et al., Reference Lee, Shih and Chang1997; Tsukimura et al., Reference Tsukimura, Bender and Linder2000), and even larvae (Yang et al., Reference Yang, Xu, Dai and Yang2005). As a precursor of vitellin (Vt), the common form of yolk proteins, vitellogenin (Vg) plays a vital role in the synthesis and secretion of yolk protein (Tufail & Takeda, Reference Tufail and Takeda2009; Guo et al., Reference Guo, Dong, Yang, Cheng, Wan, Liu, Zhou and Ye2012). The quality and quantity of the yolk proteins synthesized and accumulated in eggs are crucial for embryonic survival (Harrison, Reference Harrison1990) and development (Yang et al., Reference Yang, Xu, Dai and Yang2005). The synthesis of Vg/Vt is, therefore, considered to be a reliable indicator for evaluating female ovarian development and reproductive fitness (Tsukimura, Reference Tsukimura2001).
Reproductive regulation is essential for living organisms. This is especially critical in invasive species allowing them to achieve their reproductive potential and to successfully colonize and adapt to new environments. As a major component of oogenesis (Shapiro et al., Reference Shapiro, Wasserman, Greany and Nation2000), vitellogenesis is considered to be the core process of reproductive regulation (Guidugli et al., Reference Guidugli, Nascimento, Amdam, Barchuk, Omholt, Simões and Hartfelder2005). The reproductive success of all the oviparous animals including insects is heavily dependent on the synthesis and accumulation of Vg/Vt in oocytes. Adequate Vg/Vt production, however, requires sufficient nutrient availability, and their circulating titers are, therefore, closely associated with nutrition intake (Bitondi & Simoes, Reference Bitondi and Simoes1996). It has been suggested that nutritional condition is an important factor for reproductive success (Awmack & Leather, Reference Awmack and Leather2002; Barone & Frank, Reference Barone and Frank2003) and that Vg synthesis and protein production in the fat body can be affected by food availability (Fei et al., Reference Fei, Martin, Jaskowiak, Hatle, Whitman and Borst2005). On the other hand, the ability to rapidly expand and populate a new territory in the shortest time possible is indispensable for successful colonization by certain species especially those invasive ones. The close relationship between host plant and herbivore has been extensively studied both from the nutritional and co-evolutionary aspects (Awmack & Leather, Reference Awmack and Leather2002; Mello & Silva-Filho, Reference Mello and Silva-Filho2002; Utsumi, Reference Utsumi2011). The nutrient substances and secondary compounds inside host plants can impact the growth and development process as well as survival of herbivores, which may ultimately lead to variability of reproductive success and offspring fitness for these species (Awmack & Leather, Reference Awmack and Leather2002). Previous studies have documented the molecular characteristics (Tufail & Takeda, Reference Tufail and Takeda2005, Reference Tufail and Takeda2008; Shu et al., Reference Shu, Wang, Lu, Zhou, Zhou and Zhang2011), regulatory mechanisms (Guidugli et al., Reference Guidugli, Nascimento, Amdam, Barchuk, Omholt, Simões and Hartfelder2005; Nilsen et al., Reference Nilsen, Ihle, Frederick, Fondrk, Smedal, Hartfelder and Amdam2011), and potential roles of Vg in oogenesis (Tufail & Takeda, Reference Tufail and Takeda2005, Reference Tufail and Takeda2009) and social organization (Nelson et al., Reference Nelson, Ihle, Fondrk, Page and Amdam2007), however, few reports focus on combining the insect Vg accumulation and ovarian development or egg quality from a host nutrition-dependent aspect, especially in those invasive species with tremendous reproductive capacity. In the present study, an invasive species, the nipa palm hispid beetle, Octodonta nipae Maulik (Coleoptera: Chrysomelidae) was used to determine the effect of host plant on ovarian development. This pest is native to Malaysia (Sun et al., Reference Sun, Yu, Zhang and Wang2003) and feeds on a wide range of host plants, causing serious damage to palm trees in southeast China (Hou & Weng, Reference Hou and Weng2010). The full-length cDNA of the Vg gene in O. nipae (OnVg) was cloned and the relative expression level for different developmental stages of the beetle reared on two different host plants was also analyzed. The results determined the sex- and developmental stage-specific expression pattern of OnVg, and demonstrated the significant impact the host plant has on ovarian development and offspring quality.
Materials and methods
Insect rearing and sampling
The laboratory population of O. nipae was established from individuals collected from a nursery (25°43′42″N, 119°20′35″E) in Fuqing City, Fujian Province of China (Hou & Weng, Reference Hou and Weng2010). Two different species of host plants, Canary Island date palm Phoenix canariensis Hort. ex Chabaud and pygmy date palm P. roebelenii O’ Brien, were used as hosts to rear O. nipae over 12 generations beginning in 2012. Insect rearing was carried out under laboratory conditions (see Li et al., Reference Li, Zhang, Hou and Tang2014). O. nipae female adults fed on P. canariensis were sampled for cloning OnVg. To identify possible variations caused by selection of host plant, insects that were reared on each of the two host plants were sampled for developmental stage-specific expression analysis at different developmental stages (i.e., 4th instar larvae, 3 day-old pupae, 15 day-old virgin females and males; 15 days are needed for O. nipae to be sexually mature). Five individuals were pooled in each sample with four replications for the OnVg expression profile analysis.
Cloning the full-length cDNA of Vg gene
The Trizol reagent (Invitrogen) was used for total RNA extraction following the manufacturer's protocol. The total RNA samples were assayed for RNA concentration using the NanoDrop 2000 (Thermo Fisher Scientific Inc., Waltham MA, USA) and was subsequently stored at −80°C. The Thermo Scientific Verso cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Waltham MA, USA) was used for cDNA synthesis. One microgram of total RNA was added as template and then the Verso Enzyme Mix as reverse transcriptase and anchored Oligo dT as primers following the manufacturer's protocol.
The potential OnVg fragments were first screened from the O. nipae transcriptome database (Tang et al., Reference Tang, Chen, Hou and Meng2014). The primers were then designed by Primer3 (version 4.0.0 http://primer3.ut.ee/, see Koressaar & Remm, Reference Koressaar and Remm2007; Untergasser et al., Reference Untergasser, Cutcutache, Koressaar, Ye, Faircloth, Remm and Rozen2012). Polymerase Chain Reaction (PCR) amplifications were proceeded in a total reaction volume of 50 µl, consisting of 25 µl of 2 × Taq Plus MasterMix (Tiangen Biotech Co., Ltd., Beijing, China), 2 µl of each primer (VgF and VgR, 10 µM), 2 µl of cDNA template and 19 µl of PCR-grade water. The amplification conditions used were as follows: 94°C for 3 min, 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 5 min. The obtained PCR products were purified using a PCR purification kit (Tiangen Biotech Co., Ltd., Beijing, China). The purified DNA fragment was ligated with the pMD 18-T vector (TAKARA Biotech Co., Ltd., Dalian, China) and transformed into competent Escherichia coli cells (DH5α, Tiangen Biotech Co., Ltd., Beijing, China). The recombinants were screened for subsequent sequencing (Sangon Biotech Co., Ltd., Shanghai, China) through blue-white plaque selection on LB plates containing ampicillin (final concentration: 100 µg ml−1).
After obtaining OnVg fragments, the 3’ and 5’ rapid amplification of cDNA ends (RACE) PCRs were performed to obtain the 3’ and 5’ cDNA ends of OnVg using the SMARTer™ RACE cDNA Amplification Kit (Clontech Laboratories, Inc.). The gene-specific primers Vg3F, Vg5R, and nested primers NVg3F, NVg5R were designed for the 3’ and 5’ RACE, respectively. The cDNA template synthesis and amplification conditions were conducted according to the user manual. The subcloning and sequencing of RACE-PCR products were performed as described above except the cloning vector and competent E.coli cells used for those procedures were pEASY-T5 (TransGen Biotech Co., Ltd., Beijing. China) and Trans1-T1 Phage Resistant Chemically Competent Cell (TransGen Biotech Co., Ltd., Beijing. China), respectively. The full-length cDNA of Vg gene in O. nipae was then obtained by contig alignment of three cDNA fragments according to the overlapping region. All of the primers used for molecular cloning are available in Supplementary Table S1.
Sequence analysis
The homology of OnVg sequences with other Vgs were checked using the National Center for Biotechnology Information (NCBI) Translated BLAST tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The Expasy-Translation tool (web.expasy.org/translate/) was used to translate the full-length OnVg into the deduced amino acid sequence. The signal peptide position was detected using SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/) and the molecular weight and isoelectric point of deduced amino acid sequence were detected using the ExPASy-Compute pI/Mw tool (http://web.expasy.org/compute_pi/). The putative phosphorylation and glycosylation sites were checked through the NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/) and NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/), respectively. The NCBI Conserved Domain Search (CDD, http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used for detecting the conserved domains of the deduced amino acid sequence.
Construction of phylogenetic tree
Multiple alignments were performed using the ClustalX program (Thompson et al., Reference Thompson, Gibson, Plewniak, Jeanmougin and Higgins1997) with gap opening penalty 10, gap extension penalty 0.05, and gap separation penalty range 8. The phylogenetic tree based on the OnVg deduced amino acid sequence was constructed by MEGA 5 (Tamura et al., Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011) using the neighbor-joining (NJ) algorithm, the reliability of the clusters within the tree was tested using bootstrap resampling with 1000 replicates. The Vg sequences used for comparison and phylogenetic tree construction were available in Supplementary Table S2.
Expression profile of OnVg across different developmental stages reared on two different host plant species
Samples were collected from different developmental stages (i.e., 4th instar larvae, 3 day-old pupae, and 15 day-old virgin females and males) of O. nipae individuals fed on either P. canariensis or P. roebelenii. The total RNA extraction of the whole body was performed as described above and cDNA synthesis was achieved using the Thermo Scientific Verso cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Waltham MA, USA). SYBR Green fluorescence quantitive real-time PCR (qRT-PCR) was carried out for relative expression analysis of OnVg with ribosomal protein S3 gene (OnRPS3) being chosen as the internal reference gene. Primers qVgF and qVgR were used for OnVg amplification while RPS3F and RPS3R were used to amplify OnRPS3 (see Supplementary Table S1). The qRT-PCR was conducted in an ABI 7500 real-time PCR system and the reaction procedure consisted of the following: one cycle of 95°C for 10 min, 40 cycles of 95°C for 15 sec, 60°C for 1 min, followed by one cycle of 95°C for 15 sec, 60°C for 1 min, 95°C for 30 sec, 60°C for 15 sec (melting-curve stage for a specific PCR product verification). Each sample was assayed in triplicate and the amplification data were normalized to OnRPS3 expression. The 2−∆∆CT method was used to calculate the real-time data (Livak & Schmittgen, Reference Livak and Schmittgen2001).
Ovarian development evaluation
Females (15 day-old) fed on P. canariensis and P. roebelenii were collected for ovarian dissection. Prior to dissection, the females were anesthetized on ice and then disinfected with a 75% alcohol solution. The elytra were removed using a McPherson-Vannas scissors (World Precision Instruments, Inc., Sarasota, FL, USA). Female bodies were placed on a glass slide containing a drop of phosphate buffer saline (PBS) (NaCl 8 g, KCl 0.2 g, Na2HPO4 1.42 g, KH2PO4 0.27 g, in ultrapure water 1000 ml, pH 7.4). The insect body was cut open from the flank to the end of abdomen using the McPherson-Vannas scissors. The O. nipae female reproductive systems were isolated from the body using two Dumont forceps (World Precision Instruments, Inc., Sarasota, FL, USA). The reproductive systems were then immersed into Giemsa staining solution for 5 min followed by three washings of PBS. Each complete reproductive system was placed under a stereoscopic microscope (Nikon SMZ745 T, Japan), which connected to a digital camera (Digital Sight DS-Fi2, Nikon, Japan) for photographing. The digitalized images of the reproductive systems were then analyzed using NIS Elements D software (version 4.30, Nikon, Japan). The length, width and cross-sectional area of each O. nipae ovariole were calculated and recorded through measuring tools attached to the software.
Egg hatchability assessment
An egg hatchability study was conducted in order to evaluate the quality of eggs laid by the females. Twenty-five virgin females and an equivalent number of males fed on P. canariensis and P. roebelenii were paired and kept in a sterile transparent plastic container for mating under laboratory conditions (25 ± 0.5°C, RH: 75 ± 5%, L:D = 14:10). After 7 days, all eggs were collected and 50 eggs were added to each jar containing the corresponding host plant. The containers were kept in an incubator (25 ± 0.5°C, RH: 75 ± 5%, L:D = 14:10) for 10 days. Palm leaves of the two species were replaced daily and the newly hatched larvae were counted to determine egg hatchability. There were a total of 33 replicates of P. canariensis-fed beetles and 30 replicates of P. roebelenii-fed beetles being carried out. Among these groups, two replicates fed on P. canariensis and three replicates fed on P. roebelenii were excluded from later analysis due to the escape of newly hatched larvae.
Statistical analysis
Data were tested for normality and homogeneity of variance using the Kolmogorov–Smirnov test and Levene's test, respectively. The egg hatchability data were analyzed using an unpaired t-test. Because they did not meet the homogeneity of variance assumption, ovarian development (ovariole length, ovariole width and ovariole cross-section area) was analyzed using the Mann–Whitney test. The data for relative expression of OnVg across different developmental stages also did not confirm to homogeneity of variance, therefore, a Kruskal–Wallis test was used to examine the expression level of OnVg among developmental stages. For the expression analysis of OnVg at a particular developmental stage on each of the two host plants, the data for the pupal and male adult stages were analyzed using an unpaired t-test while the larval and female adult stage data were analyzed using the Mann–Whitney test due to their heteroschedasticity. In addition, the linear regression analysis was used to test the potential correlation between OnVg expression and egg hatchability. To minimize the impact of sampling on correlation analysis, females from the same batch were collected and then divided into two groups, one group was used for the expression analysis of OnVg and the other one for the egg hatchability assay. All statistical analyses were conducted using SPSS, version 17.0 (SPSS Inc., Chicago, Illinois). The figures were mapped using SigmaPlot for Windows version 12.0 (Systat Software Inc., San Jose, California).
Results
Ovarian development assessment and egg hatchability
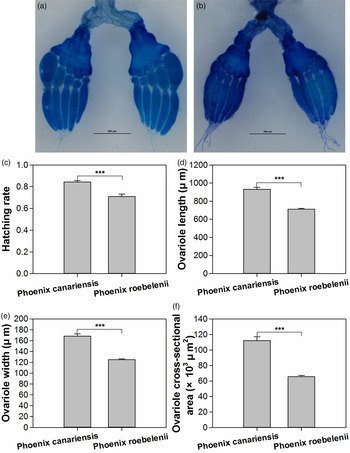
The host plants significantly influenced the ovarian development of female O. nipae (fig. 1a, b). Females fed on P. canariensis showed significantly higher ovariole length (Mann–Whitney test: Z = −7.429, P < 0.001), width (Mann–Whitney test: Z = −8.355, P < 0.001) and cross-sectional area (Mann–Whitney test: Z = −7.918, P < 0.001) than those fed on P. roebelenii (fig. 1d–f).

Fig. 1. Ovarian dissection of female Octodonta nipae fed on P. canariensis (a) and P. roebelenii (b), and effects of host plants on the egg hatchability (c), ovariole length (d), ovariole width (e) and ovariole cross-sectional area (f). Data were shown as Mean ± SE. Asterisks above the bars indicate statistically significant difference (***P < 0.001).
Moreover, the hatchability of eggs laid by females fed on different host plants was significantly different (unpaired t-test, t 1,52 = 4.935, P < 0.001). The hatch rate of eggs laid by females fed on P. canariensis (84.29 ± 1.48%) was significantly higher than that of females fed on P. roebelenii (70.92 ± 2.32%) (fig. 1c).
Analysis of the full-length cDNA of Vg gene (OnVg)
The complete OnVg was 5451-bp long, including a 12 bp 5’-untranslated region (UTR), a 69 bp 3’-UTR and a 5370 bp opening reading frame (ORF), which encoded 1789 amino acid residues (accession number: KR736347 and Supplementary Figure S1). The first 18 amino acid residues of ORF were predicted signal peptide through the SignalP program (Supplementary Figure S1). The predicted molecular weight and isoelectric point of the mature protein were 204.27 kDa and 4.72, respectively. A Vg-N domain (amino acid position: 22–717) with a gap (amino acid position: 325–387), a DUF 1943 domain (amino acid position: 774–1042) and a von Willebrand factor type D domain (amino acid position: 1461–1645), were identified using a NCBI CCD search. In addition, six potential cleavage recognition sites RXXR and two conserved amino acid motifs GL/ICG (here GICG) and DGXR (here DGQR) were identified. Additionally, the putative glycosylation site (NXS/T) and potential phosphorylation residues were also found in the Vg sequence in O. nipae (Supplementary Figure S1).
A NJ tree construction was used to illustrate the evolutionary relationship among different Vgs (fig. 2). The dendrogram showed that the OnVg was placed with other insects especially other Coleoptera Vgs (i.e., Anthonomus grandis, Tenebrio molitor, Tribolium castaneum) into a single cluster. It also indicated that the coleopteran Vgs appear to have a closer evolutionary relationship with the hymenopteran Vgs than with the Vgs of other insects. The phylogenetic tree divided the Vgs from different orders into separate clusters reflecting their phylogeny. Moreover, the dendrogram clustered different insect orders into one homogeneous group, showing the existence of a greater sequence similarity that extends beyond the insect order level. Additionally, it seems to imply that the hymenopteran Vgs are the sister group of coleopteran Vgs while the dipteran Vgs are the sister group of hemipteran Vgs. However, the lepidopteran Vgs showed a distant phylogenetic relationship with other insect Vgs.

Fig. 2. Molecular phylogenetic tree comparing the deduced amino acid sequences of Octodonta nipae Vg with other Vgs. The phylogenetic analysis was initially performed using the ClustalX program for multiple sequence alignment and followed by MEGA5 for a neighbor-joining tree construction. The numbers at each node are the percentage of 1000 bootstrap resampling.
Expression profile of OnVg
The OnVg was specifically expressed in female adults regardless of host plant (P. canariensis: Kruskal–Wallis test, d.f. = 3, χ2 = 8.824, P = 0.032; P. roebelenii: Kruskal–Wallis test, d.f. = 3, χ2 = 8.500, P = 0.037). OnVg expression was barely detected in the larvae, pupae and male adults (fig. 3). In addition, no significant differences in terms of OnVg expression were found in the larval (Mann–Whitney test: Z = 0.00, P = 1.00), pupal (unpaired t-test: t 1,6 = 0.214, P = 0.838) and male adult stages (unpaired t-test: t 1,6 = −0.338, P = 0.747). However, the OnVg expression in the female adult stage was significantly affected by host plants (Mann–Whitney test: Z = −2.309, P = 0.021). Females fed on P. canariensis showed a much higher OnVg expression level than those fed on P. roebelenii (fig. 3).

Fig. 3. Relative expression level of OnVg in different developmental stages fed on two different host plants. Asterisks above the bars indicate statistically significant difference (*P < 0.05), NS above the columns denote non-significant difference. Data were shown as Mean ± SE.
Relationship between expression profile of OnVg and offspring quality
Regression analysis was used to illuminate the potential relevance between OnVg expression and offspring quality. Despite the host plant, a significant positive correlation between OnVg expression and egg hatchability was detected in females fed on either P. canariensis (F 1,29 = 56.836, P < 0.001, fig. 4) or P. roebelenii (F 1,25 = 67.820, P < 0.001, fig. 4). Those females with a higher OnVg expression level were inclined to lay higher-quality eggs (with high egg hatching rates).

Fig. 4. Regression analysis between OnVg expression level and offspring quality. Females from the same batch were used for the gene expression analysis and the egg hatchability assay.
Discussion
Numerous Vg cDNAs including several complete Vg cDNAs have been sequenced in hymenopteran insects (Pinto et al., Reference Pinto, Bitondi and Simões2000; Guidugli et al., Reference Guidugli, Nascimento, Amdam, Barchuk, Omholt, Simões and Hartfelder2005; Nelson et al., Reference Nelson, Ihle, Fondrk, Page and Amdam2007), dipteran insects (Ahmed et al., Reference Ahmed, Maingon, Romans and Hurd2001; Dana et al., Reference Dana, Hillenmeyer, Lobo, Kern, Romans and Collins2006), hemipteran insects (Lee et al., Reference Lee, Hatakeyama and Oishi2000; Guo et al., Reference Guo, Dong, Yang, Cheng, Wan, Liu, Zhou and Ye2012), dictyopteran insects (Tufail & Takeda, Reference Tufail and Takeda2005; Ciudad et al., Reference Ciudad, Piulachs and Bellés2006) and lepidopteran insects (Meng et al., Reference Meng, Liu, Shiomi, Nakagaki and Kajiura2008; Shu et al., Reference Shu, Wang, Lu, Zhou, Zhou and Zhang2011) while only a limited number of Vgs in coleopteran insects have been reported. In the present study, the full-length Vg in O. nipae was cloned and its evolutionary relationship with other Vgs was investigated. The deduced amino acid sequence of OnVg contained typical domains and conserved motifs that are commonly found in insect Vg. In addition, a close molecular evolutionary relationship in the sequence similarity of Vg genes between O. nipae and its subordinative insect order was also verified through multiple alignment. Furthermore, our results showing the close relationship between Vg expression level and offspring quality should be applicable to many pest management situations through reproductive regulation in insects especially those invasive ones.
In line with previous studies (Lee et al., Reference Lee, Shih and Chang1997; Dong et al., Reference Dong, Ye, Yao, Huang, Chen, Shen and Hu2008), the results in this study demonstrate the characteristic female-specific expression of Vg. The specific expression of OnVg in the female adult stage also suggests a developmental stage-specific expression pattern of Vg in O. nipae, which may be attributable to the variation in reproductive physiology resulting from peculiarities in an individual's life history. In most insect species, the degree of ovarian maturation directly contributes to the oviposition and egg quality (Papaj, Reference Papaj2000). Since nutrition both in the larval and adult stages is important for the realization of reproductive fitness, and feeding is an obvious prerequisite of egg production, food quality and availability would strongly influence reproductive capability. Therefore, the reproductive efficiency of herbivores would be expected to decrease when fed on low host quality or availability and vice versa. In the present study, P. canariensis has a higher crude fat, soluble protein, and free amino acid content than P. roebelenii (data not shown here), which coincides with the above prediction. Oviposition is a dynamic consequence in which the female individual in respond to the variation of host quality and availability from a functional way (Papaj, Reference Papaj2000). Egg production is therefore generally considered to be a reliable indicator of female reproductive fitness in response to differing potential factors (e.g., abiotic factors such as temperature and humidity or biotic factors such as host quality and prey availability, etc.). On the other hand, dissection of the reproductive system provides a direct and visual assessment of ovarian development from the aspect of developmental biology. In the present study, both the higher egg hatchability and the more developed reproductive system found in female O. nipae fed on P. canariensis indicated a higher offspring fitness and an increased female reproductive efficiency, which corresponds to the beetle's host preference (O. nipae prefers to feed on P. canariensis rather than P. roebelenii, personal observation). Finally, being the most important nutritional source, Vg provides the growing oocytes, embryos, and even larvae with various nutrients (see the section Introduction). The significant correlation between the OnVg expression level and ovarian development in the present study provides us with robust evidence regarding the promotion of Vg accumulation on reproductive success.
It has been suggested that Vg has comprehensive regulatory functions and relates to some life history traits in honeybees (Guidugli et al., Reference Guidugli, Nascimento, Amdam, Barchuk, Omholt, Simões and Hartfelder2005; Nelson et al., Reference Nelson, Ihle, Fondrk, Page and Amdam2007). The pleiotropic effects of the Vg gene on various physiological processes have also been demonstrated in honeybees (Nelson et al., Reference Nelson, Ihle, Fondrk, Page and Amdam2007). However, in most insects, hormonal regulation especially juvenile hormone (JH) is an important endocrine and metabolic regulator in vitellogenesis, including Vg biosynthesis and its absorption and translocation into growing oocytes (Raikhel et al., Reference Raikhel, Brown, Bellés, Gilbert, Iatrou and Gill2005; Dong et al., Reference Dong, Ye, Yao, Huang, Chen, Shen and Hu2008). For example, JH inhibits the Vg synthesis and accumulation in honeybees when treated with high doses of pyriproxyfen (a potent JH analogue) (Pinto et al., Reference Pinto, Bitondi and Simões2000), and the hemolymph titer of JH is sensitive to life stage activities such as foraging and reproduction (Robinson et al., Reference Robinson, Strambi, Strambi and Huang1992). Besides, in response to the accumulation of ecdysteroids, the expression of Vg, vitellogenic carboxypeptidase and vitellogenic cathepsin B genes are increased during vitellogenesis in Aedes aegypti (Raikhel et al., Reference Raikhel, Brown, Bellés, Gilbert, Iatrou and Gill2005). In the present study, host plants significantly affect the Vg expression level and ovarian development. Despite the factors of hormones and host plant nutrition influencing the Vg gene expression, the presence of the insulin/insulin-like signaling (IIS) pathway (Corona et al., Reference Corona, Velarde, Remolina, Moran-Lauter, Wang, Hughes and Robinson2007) and the target of rapamycin (TOR) pathway (Patel et al., Reference Patel, Fondrk, Kaftanoglu, Emore, Hunt, Frederick and Amdam2007) drive the complex relationship that exists between nutrition status, Vg expression, hormonal regulation and social behavior, and are responsible for even more complex interactions in honeybees (Nilsen et al., Reference Nilsen, Ihle, Frederick, Fondrk, Smedal, Hartfelder and Amdam2011). For example, some key components of IIS pathway such as insulin-like peptides, insulin-like receptors and transcription factor FOXO have been reported to play a part in Vg synthesis by a feedback loop mechanism (Corona et al., Reference Corona, Velarde, Remolina, Moran-Lauter, Wang, Hughes and Robinson2007). And a positive effect of TOR on Vg expression has been verified in both honeybees (Patel et al., Reference Patel, Fondrk, Kaftanoglu, Emore, Hunt, Frederick and Amdam2007) and mosquitoes (Hansen et al., Reference Hansen, Attardo, Park, Peng and Raikhel2004). Therefore, the functional verification of OnVg on ovarian development by means of RNA interference and the correlation between hormones and Vg expression and their interaction with other nutrient-sensitive target genes are necessary in the future for a broad understanding of insect Vg.
In summary, the present study clones the complete Vg gene in O. nipae, analyzes the expression profile of OnVg and verifies its correlation with ovarian development and subsequent egg quality, enhancing our understanding regarding Vg expression levels and the effects of differing host plant environments on ovarian development. In addition, data from this study may also have practical applications in pest management programs through the use of artificial reproductive disturbance by alternating host plant species or varieties or by reproductive regulation through vitellogenesis mediated by specific endocrine hormones.
Supplementary Material
The supplementary material for this article can be found at http://dx.doi.org/10.1017/S0007485316000353.
Acknowledgements
We gratefully thank Xiaoqiang Yu (University of Missouri-Kansas City) and Cecil L. Smith (Georgia Museum of Natural History) for their critical comments on the manuscript. This work was supported by the National Natural Science Foundation of China (31272108, 31471829).
Conflict of Interest
The authors declare that they have no conflict of interest.