Introduction
As shown in many cases of insect chemosensory systems, olfaction play critical roles in the interactions of insects with their environment, such as foraging, oviposition behaviors, mating choice and the communication of social insects (Hallem et al., Reference Hallem, Dahanukar and Carlson2006; Carey et al., Reference Carey, Wang, Su, Zwiebel and Carlson2010; Zheng et al., Reference Zheng, Peng, Zhu, Zhang, Saccone and Zhang2013). Various chemosensory genes are involved in the capture of volatiles from environment and signal transduction, including odorant-binding proteins (OBPs), chemosensory proteins (CSPs), sensory neuron membrane proteins (SNMPs), odorant receptors (ORs), gustatory receptors (GRs) and ionotropic receptors (IRs) (Hallem et al., Reference Hallem, Dahanukar and Carlson2006; Kang et al., Reference Kang, Tian, Liu, Liu, Jing and Liu2017a). In general, odors in environment were recognized and bound by two classes of small, water-soluble, extracellular-binding proteins, OBPs and CSPs, which transferred odorants through the sensilla lymph to insect chemoreceptors at the membrane surface of olfactory sensory neurons (OSNs), that were thought to be the first biochemical step in odors reception (Zheng et al., Reference Zheng, Peng, Zhu, Zhang, Saccone and Zhang2013; Chang et al., Reference Chang, Liu, Yang, Pelosi, Dong and Wang2015; Northey et al., Reference Northey, Venthur, De Biasio, Chauviac, Cole, Ribeiro, Grossi, Falabella, Field, Keep and Zhou2016).
The functions of insect OBPs have been proposed: (1) transporting hydrophobic odorants across the sensilla lymph to the chemoreceptors; (2) solubilizing hydrophobic odors, (3) concentrating odors in the sensilla lymph fluid; (4) combine with odors degrading enzymes to remove or deactivate odorants after simulating the chemoreceptors (Gu et al., Reference Gu, Zhou, Gao, Wang, Li, Guo and Zhang2015; Zhu et al., Reference Zhu, Ban, Song, Liu, Pelosi and Wang2016). In aphid, the systemic identification of OBPs only reported in the pea aphid Acyrthosiphon pisum, cotton aphid Aphis gossypii and the English grain aphid, Sitobion avenae (Fabricius) (He et al., Reference He, Zhang, Liu, Zhang, Yang and Dong2011; Zhou et al., Reference Zhou, Vieira, He, Smadja, Liu, Rozas and Field2010; Zhou et al., Reference Zhou, Min, Tang and Wang2015; Xue et al., Reference Xue, Fan, Zhang, Xu, Han, Sun and Chen2016). In Megoura viciae and Nasonovia ribisnigri, only the aphid pheromone (E-beta-farnesene) binding protein, OBP3, was cloned and functionally analyzed which was similar with the OBP3 in A. pisum (Qiao et al., Reference Qiao, Tuccori, He, Gazzano, Field, Zhou and Pelosi2009; Northey et al., Reference Northey, Venthur, De Biasio, Chauviac, Cole, Ribeiro, Grossi, Falabella, Field, Keep and Zhou2016). Consist with the results of OBP3 in A. pisum, OBP7 in the wheat aphid S. avenae showed high binding affinity with (E)-beta-farnesene (Zhong et al., Reference Zhong, Yin, Deng, Li and Cao2012). Similarly to OBPs, CSPs also mainly functioned in olfaction by solubilizing and transporting odors (Wang et al., Reference Wang, Li, Min, Mi, Zhou and Wang2014; Cui et al., Reference Cui, Gu, Zhu, Wei, Liu, Khalid, Guo and Zhang2017). However, in the vetch aphid Megoura viciae, two CSPs MvOS-D1 and MvOS-D2 showed no binding affinity for any of 28 compounds (Jacobs et al., Reference Jacobs, Liggins, Zhou, Pickett, Jin and Field2005). To consistent with this result, in Camponotus japonicas and Chrysoperla sinica, some CSPs are highly expressed in the non-chemosensory organs in the carpenter ant (Hojo et al., Reference Hojo, Ishii, Sakura, Yamaguchi, Shigenobu and Ozaki2015; Li et al., Reference Li, Zhang, Luo, Wang, Wang, Lv, Dong and Cui2015c). Thus, all these results suggested that CSPs may have varied functions in other biological function process. Additionally, SNMPs has been proposed to play an essential role in insect olfaction, especially in the process of pheromone reception in insects (Liu et al., Reference Liu, Zhang, Liu, Wang and Dong2014; Jiang et al., Reference Jiang, Pregitzer, Grosse-Wilde, Breer and Krieger2016).
Chemoreceptors, including three large and divergent families: ORs, GRs and IRs, are responsible for receiving, transporting and triggering responses to semiochemicals (Hallem et al., Reference Hallem, Dahanukar and Carlson2006; Li et al., Reference Li, Zhu, Wang, Wang, He, Chen, Sun, Deng and Zhang2015b; Liu et al., Reference Liu, Rao, Li, Feng, He and Li2015). In insect, chemoreceptors have been identified in various insect species (Ahmed et al., Reference Ahmed, Zhang, Wang, He and Bai2016; Niu et al., Reference Niu, Liu, Dong and Dong2016; Sheng et al., Reference Sheng, Liao, Zheng, Zhou, Xu, Song, He, Zhang and Wu2017). In aphid, chemoreceptors have been thoroughly identified in A. pisum, D. noxia and A. gossypii (Cao et al., Reference Cao, Liu, Walker, Li and Wang2014; Nicholson et al., Reference Nicholson, Nickerson, Dean, Song, Hoyt, Rhee, Kim and Puterka2015). Based on the real-time quantitative polymerase chain reaction (RT-qPCR) results of A. gossypii, almost all of the putative ORs were mainly expressed in the head (Cao et al., Reference Cao, Liu, Walker, Li and Wang2014). Based on the RNAi experiment, SaveOrco was considered to play a critical role in the aphid's response to pheromones and other volatiles in S. avenae (Fan et al., Reference Fan, Zhang, Francis, Cheng, Sun and Chen2015). In the Chinese white pine beetle Dendroctonus armandi, the silencing of Orco led to significantly declining in EAG response to 11 major volatiles of its host (Zhang et al., Reference Zhang, Gao and Chen2016a). As ORs serve as the molecular interface between the insect and its odor environment, GRs were considered to play a central role in co-ordinating insect feeding behaviors (Agnihotri et al., Reference Agnihotri, Roy and Joshi2016; Xu et al., Reference Xu, Papanicolaou, Zhang and Anderson2016). In vitro studies with GRs from the silkworm Bombyx mori, BmGR8 and BmGR9 showed strong responses to myo-inositol and D-fructose (Sato et al., Reference Sato, Tanaka and Touhara2011). In the cotton bollworm Helicoverpa armigera, HaGr1 and HaGr3 were indispensable and sufficient for CO2 sensing in labial palps (Ning et al., Reference Ning, Yang, Xu, Huang and Wang2016). As the third type of chemoreceptors, IR has facilitated dissection of the mechanism by which these olfactory receptors localize to OSN sensory cilia, recognize odors and produce neuronal depolarization (Wang et al., Reference Wang, Yang, Chen, Jiang, Li, Wang and Kang2015b; Hussain et al., Reference Hussain, Zhang, Ucpunar, Svensson, Quillery, Gompel, Ignell and Grunwald Kadow2016). For example, the co-expression of Ir8a was necessary for the successful expression of Ir84a in Or22a neurons or in Xenopus oocytes (Abuin et al., Reference Abuin, Bargeton, Ulbrich, Isacoff, Kellenberger and Benton2011). And co-expression of Ir8a with Ir75a conferred responses to a different ligand, propionic acid (Ai et al., Reference Ai, Min, Grosjean, Leblanc, Bell, Benton and Suh2010). By contrast, in Locusta migratoria, the IR-based pathway was not responsible for the attractive behavior of gregarious nymphs (Wang et al., Reference Wang, Yang, Chen, Jiang, Li, Wang and Kang2015b). All of these work suggested that chemosensory genes play a critical role in insect-environment interactions.
The bird cherry-oat aphid, Rhopalosiphum padi (L.), is well known as one of the most important pests of wheat. Due to direct feeding and transmitting the Barley yellow dwarf virus (BYDV), this aphid caused a dramatic reduction of the quality and yield of wheat crops globally (Li et al., Reference Li, Zhu, Wang, Wang, He, Chen, Sun, Deng and Zhang2015b; Wang et al., Reference Wang, Wu, Liu, Wu and Wang2015a). However, R. padi is a polyphagous pest of barley, oats, wheat, common choke-cherry, bird cherry Prunus padus L. and other species with alternation of hosts: its winter hosts are Rosaceae, and its summer hosts are Gramineae (Dixon, Reference Dixon1971; Li et al., Reference Li, Zhu, Wang, Wang, He, Chen, Sun, Deng and Zhang2015b). It has been found that the volatile components emitted from its host plant cause the switch between winter and summer hosts (Pettersson et al., Reference Pettersson, Pickett, Pye, Quiroz, Smart, Wadhams and Woodcock1994). Methyl salicylate produced by P. padus has been identified as a take-off stimulus of spring migrants and significantly reduced the initial aphid settling in filed spraying experiment (Park et al., Reference Park, Elias, Donato and Hardie2000; Pettersson et al., Reference Pettersson, Pickett, Pye, Quiroz, Smart, Wadhams and Woodcock1994). Besides, the nonvolatile components of its host plants play a critical role in the host acceptance (Dewhirst & Pickett, Reference Dewhirst and Pickett2009; Nam & Hardie, Reference Nam and Hardie2014). And the feeding behaviors of R. padi in different hosts were significantly different (Nam & Hardie, Reference Nam and Hardie2014; Li et al., Reference Li, Zhu, Wang, Wang, He, Chen, Sun, Deng and Zhang2015b). All of these results revealed that R. padi has evolved comprehensive chemosensory systems to search for the suitable host plants during their migration and alter their feeding strategy based on the host plants’ nutrition and defense condition. In this study, we identified and annotated the putative chemosensory genes in R. padi using next-generation sequencing. To have an initial prediction of these genes function, the expression patterns of these genes among the different tissues, hosts and wing morphs were conducted by qPCR.
Materials and methods
Identification of the chemosensory genes in R. padi
The transcriptome data were downloaded from the National Center for Biotechnology Information (NCBI) Sequences Read Archive (SRA) database (Accession number: ERR983159, ERR983160, ERR983161, ERR983162, ERR983163 and ERR983164), which were conducted by Illumina HiSeq 2000 paired-end sequencing. The raw reads were cleaned by removing adapter sequences, low-quality sequences (reads with ambiguous bases ‘N’), and reads with >50% Q <20 bases. Cleaned reads shorter than 60 bases were removed because short reads might represent sequencing artifacts. Then, transcriptome de novo assembly was carried out using a short-reads assembling program – Trinity, which combines three independent software modules: Inchworm, Chrysalis and Butterfly, to overcome the quality and polymorphism issues. The previously described OBPs, CSPs, SNMPs, ORs, GRs and IRs sequences from A. pisum, A. gossypii and D. melanogaster were used as queries searching and verifying against the R. padi database using TBLASTX program with e-value <10−5.
Phylogeny analysis of the chemosensory genes
Amino acid sequences of candidate OBPs, CSPs, SNMPs, ORs, GRs or IRs were aligned by MAFFT through FFT-NS-I iterative refinement method with JTT200 scoring matrix, unalignlevel 0.3, ‘leave gappy regions’ set and other default parameters. Bioedit Sequence Alignment Editor 7.1.3.0 (Ibis Pharmaceuticals, Inc., Carlsbad, CA, USA) was used for the further manual editing. Phylogenetic trees were subsequently constructed by the Maximum likelihood (ML) method using PhyML3.1 based on the best-fit model LG+G estimated by ProtTest2.4. SH-like approximate likelihood ratios (aLRT-SH) supports were used to evaluate the reliability of internal branches. The trees were further edited using the ITOL tool. And all amino acid sequences used in this work were presented in table S2.
The rear condition and treatment of R. padi
In this study, the bird-cherry oat, R. padi was kindly provided by Dr Kang Wang (Northwest A&F University, Yangling, Shaanxi, China), and maintained on seedlings of wheat cultivar ‘Xinong 979’ at 23 ± 1°C, a photoperiod of L16:D8, and relative humidity of 60 ± 5%. For the host switch treatment, the third nymph of R. padi was transferred from wheat to maize (Zea mays L., var. ‘Zhengdan 985’) and chili pepper (Capsicum annuum L., var. ‘Lingxiudajiao F1’) upon to adult. The whole bodies of five R. padi from different treatment were collected as a biological replicate and each treatment with three biological treatments. Then, the collected samples were frozen in liquid nitrogen and immediately stored at −80°C for further processes.
Expression profiles of chemosensory genes via qPCR
Total RNA was extracted by TRIzol reagent (Takara Bio, Tokyo, Japan) per manufacturer's instructions. The RNA integrity was verified by 1% agarose gel electrophoresis and the quantity was assessed with a Nanodrop ND-2000 spectrophotometer.
qPCR was performed to validate the expression of candidate chemosensory genes in R. padi. Then, 1 µg of RNA was used to synthesize the first strand complementary DNA (cDNA) using a PrimeScript® RT reagent Kit with gDNA Eraser (perfect Real Time) (Takara, Tokyo, Japan) following the manufacturer's protocol. The synthesized cDNA was stored at −20°C. Gene-specific primers were designed by Primer Premier 5 (PREMIER Biosoft International, Palo Alto, CA, USA) and were shown in table S1. qPCR was conducted in 20 µl reactions containing 50x SYBR Premix Ex Taq 10 µl, primer (10 mM) 0.8 µl, sample cDNA 0.8 µl, and sterilized ultra-pure grade H2O 7.6 µl. Cycling conditions were: 95°C for 30 s, 40 cycles of 95°C for 5 s and 55°C for 30 s. Each sample was done in triplicate technical replicates and three biological replicates. Relative quantification was performed by using the Comparative 2−ΔΔCT method. Transcription levels of these genes were normalized by Actin (Zuo et al., Reference Zuo, Peng, Wang, Lin, Li and Chen2016; Kang et al., Reference Kang, Liu, Tian, Zhang, Guo and Liu2017b).
Results
Identification of putative OBPs
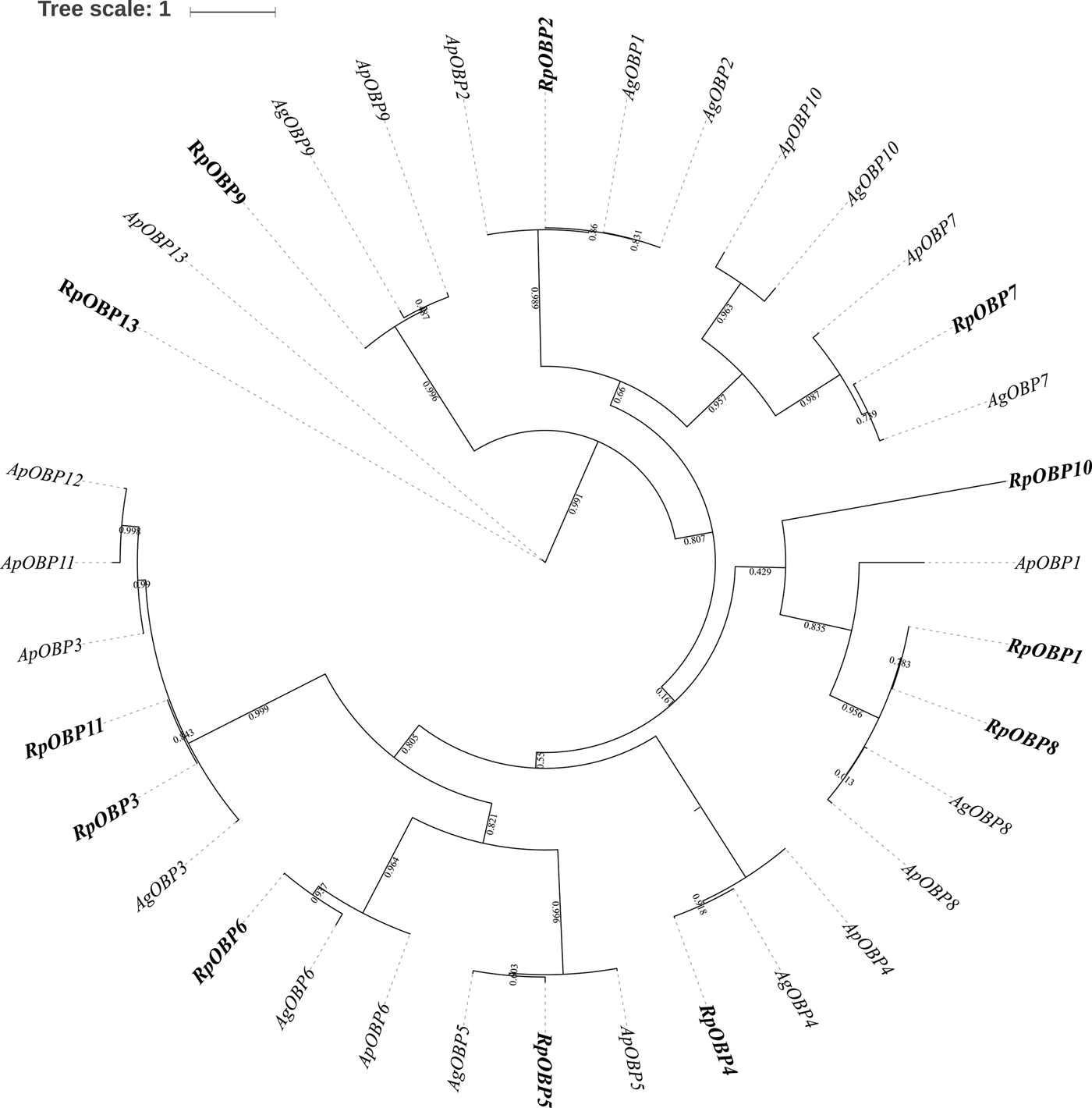
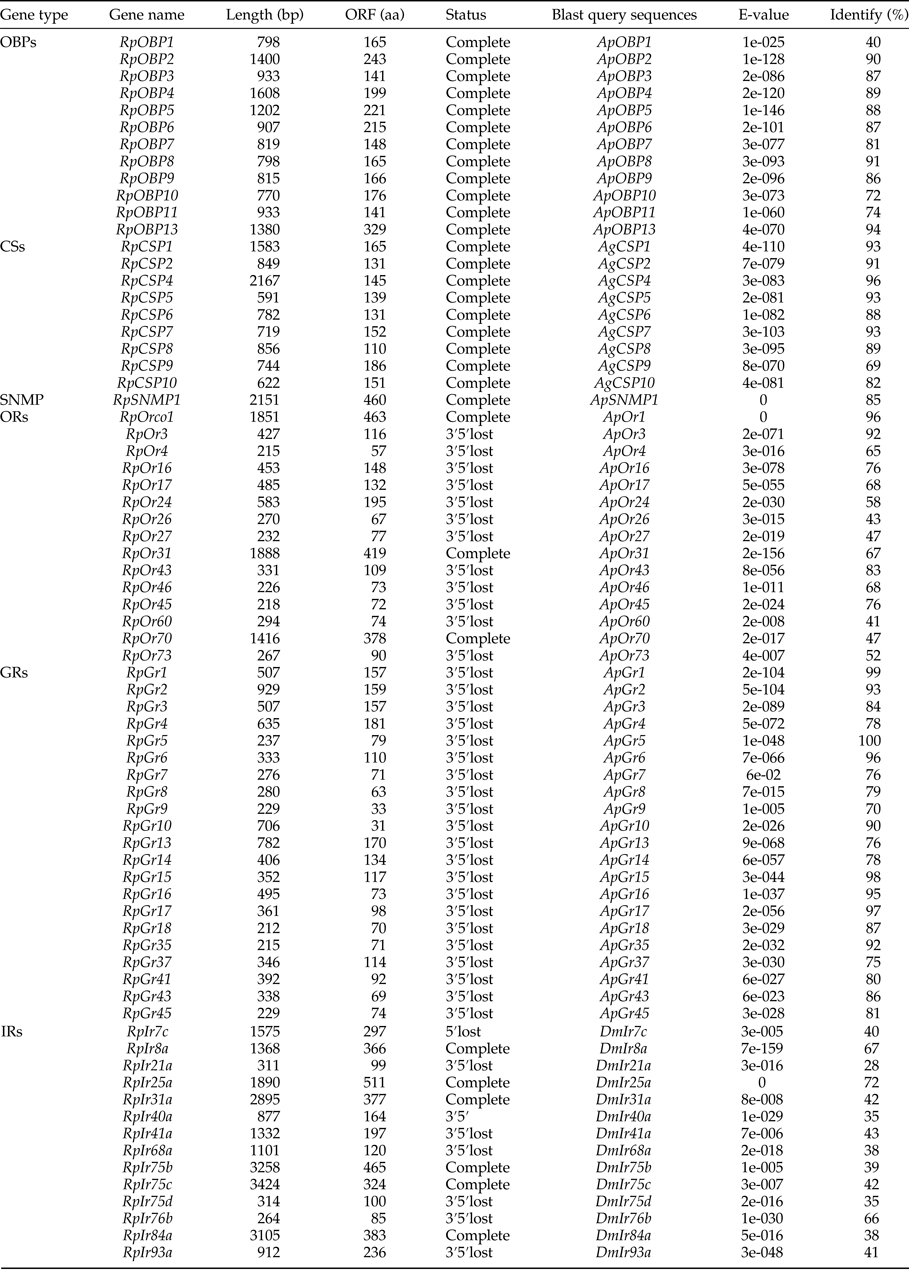
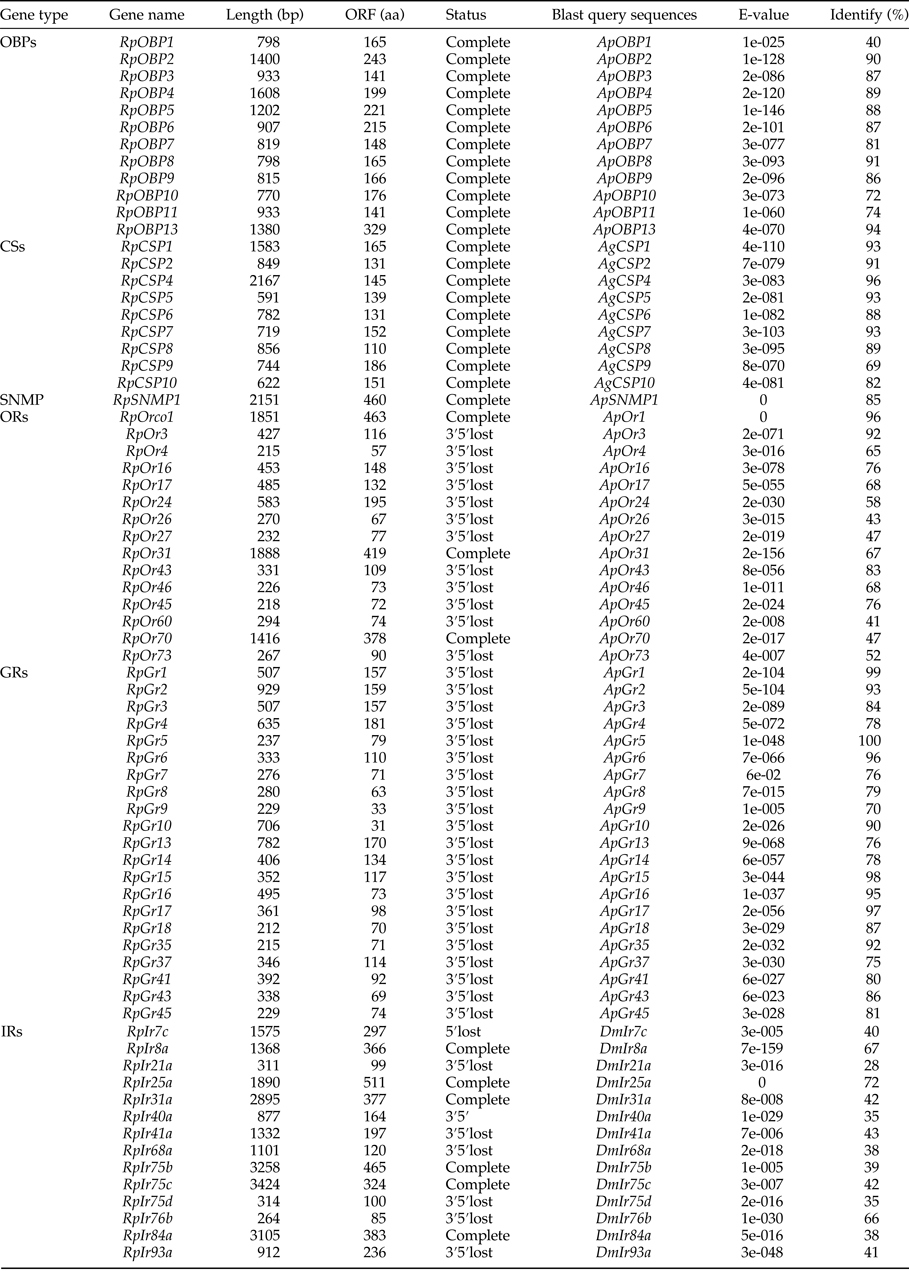
Fourteen transcripts encoding candidate OBPs in R. padi were identified based on the PSI-blast. The number of putative OBPs in this work is more than that of A. gossypii and A. pisum. All of these putative OBPs had full-length ORFs, and only RpOBP10 without signal peptide. The detail information on the putative OBPs was shown in table 1. And a phylogenetic tree was constructed using the identified OBPs from A. pisum, A. gosyypii and R. padi (fig. 1). In the phylogenetic tree, RpOBP2, RpOBP4, RpOBP5, RpOBP6, RpOBP8, RpOBP7 and RpOBP9 were clustered in a specific subgroup.

Fig. 1. Maximum likelihood phylogenetic tree of odorant-binding proteins (OBPs). OBPs from R. padi (Rp), A. pisum (Ap) and A. gossypii (Ag) are included.
Table 1. The identified chemosensory genes in R. padi transcriptome
Identification of putative CSPs
Nine transcripts encoding candidate CSPs in R. padi were identified and the detail information on the putative CSPs was shown in table 1. The number of putative CSPs in R. padi is similar with A. gossypii and A. pisum and more than that of S. avenae. Of the nine putative CSPs, all of them had full-length ORFs and only RpCSP8 without signal peptide. To analyze the relationship between the CSPs and those of other species, a phylogenetic tree was presented in fig. S1, which includes the identified OBPs from R. padi, A. gossypii and N. lugens. In the phylogenetic tree, all of these identified CSPs were clustered in a specific subgroup.
Identification of putative SNMPs
Only one transcript that encoded putative SNMP with ORFs of 1470 bp (table 1). The E-value for Blastp search was 0, indicating that they were homologous to known sequences in A. pisum and Diuraphis noxia. The phylogenetic tree was constructed (fig. S2).
Identification of putative ORs
We identified transcripts encoding 14 putative ORs. Of the 14 ORs, RpOrco1, RpOr17 and RpOr31 likely represented full-length genes, encoding proteins of longer than 300 amino acids. And the E-value for Blastp search of RpOrco1 was 0 comparing with that of A. pisum and A. gossypii. The detail information on ORs was shown in table 1, and phylogenetic tree was constructed and presented in fig. 2. RpOrco1, AgOrco1 and ApOr1 were clustered in a specific subgroup called odorant co-receptor (Orco).

Fig. 2. Maximum likelihood phylogenetic tree of odorant receptors (ORs, A) and the amino acid sequences alignment of odorant co-receptors (Orco) of R. padi (B). ORs from R. padi (Rp), A. pisum (Ap) and A. gossypii (Ag) are included.
Identification of putative GRs
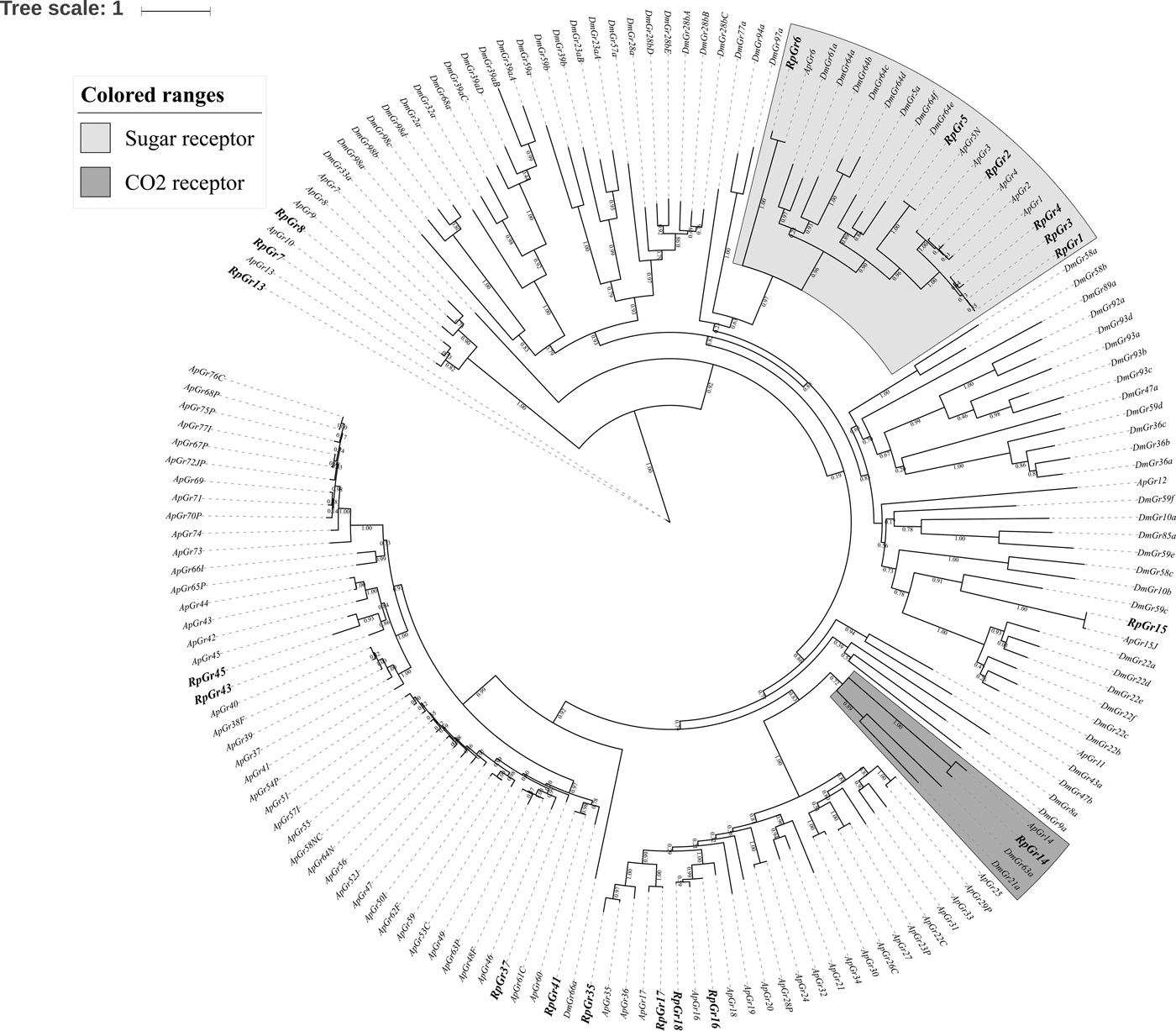
We identified 19 transcripts encoding putative GRs (table 1). However, none of them were close to full-length genes with bigger than 1000 bp ORFs. A phylogenetic tree was constructed with sequences from R. padi, A. pisum and D. melanogater. In the phylogenetic tree, RpGr14 was clustered as carbon dioxide receptor (CO2 receptor), and RpGr1, RpGr2, RpGr3, RpGr4 and RpGr6 were found in a clade with sugar receptors, which included GRs identified from D. melanogater (fig. 3).

Fig. 3. Maximum likelihood phylogenetic tree of gustatory receptors (GRs). GRs from R. padi (Rp), A. pisum (Ap) and D. melanogaster (Dm) are included.
Identification of putative IRs
We identified 16 transcripts encoding putative IRs (table 1). Among them, 12 IRs were close to the full-length gene. The E-value of RpIr25a was 0 comparing with the sequence of Ir25a in Drosophila melanogaster. In the phylogenetic tree, most of these IRs were clustered as a known group, like Ir8a, Ir25a, Nmdar, Ir93a, Ir75b and Ir40a (fig. 4).

Fig. 4. Maximum likelihood phylogenetic tree of ionotropic receptors (IRs). IRs from R. padi (Rp), A. pisum (Ap), D. melanogaster (Dm) and A. gossypii (Ag) are included.
The expression patterns of putative chemosensory genes
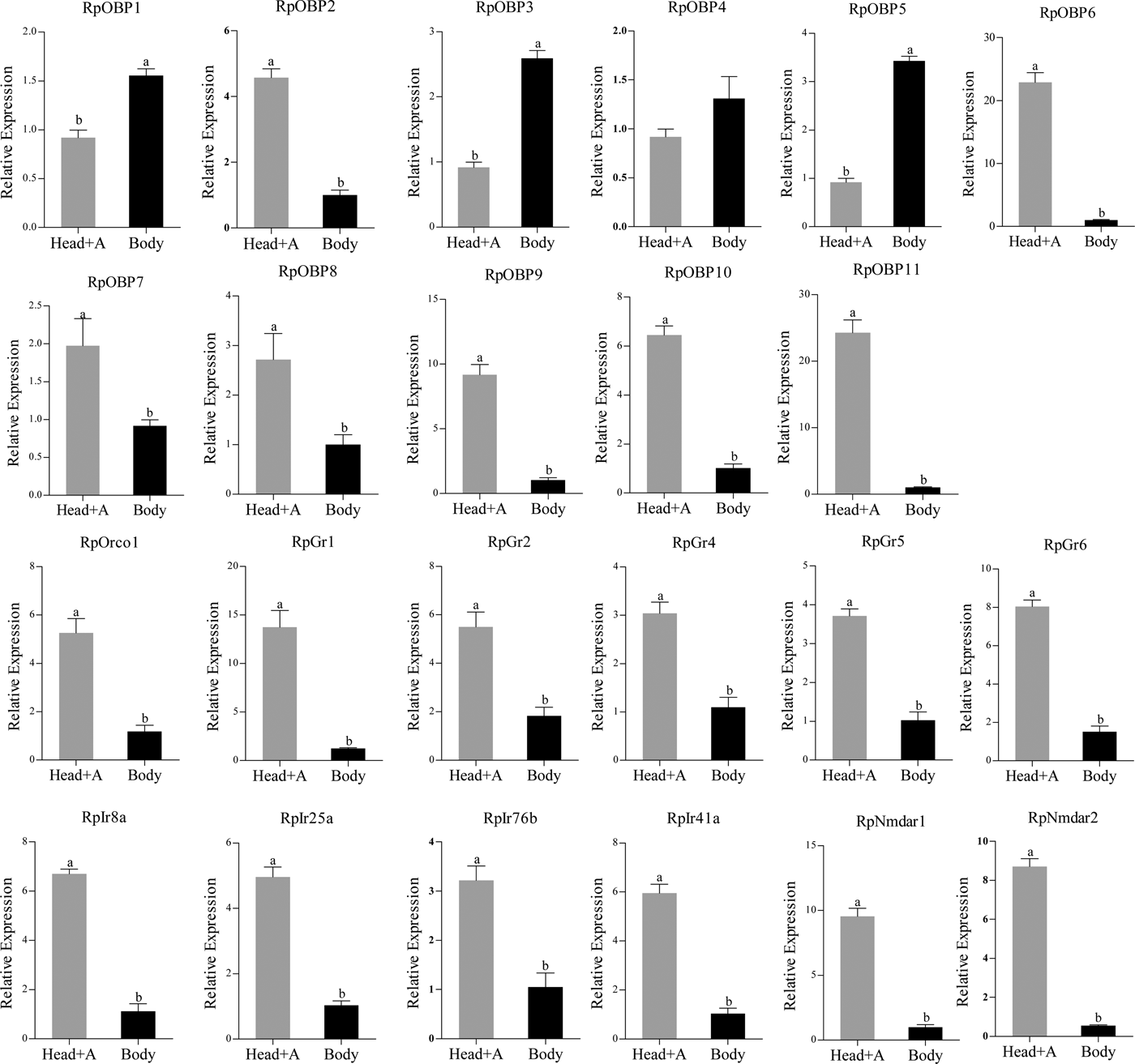
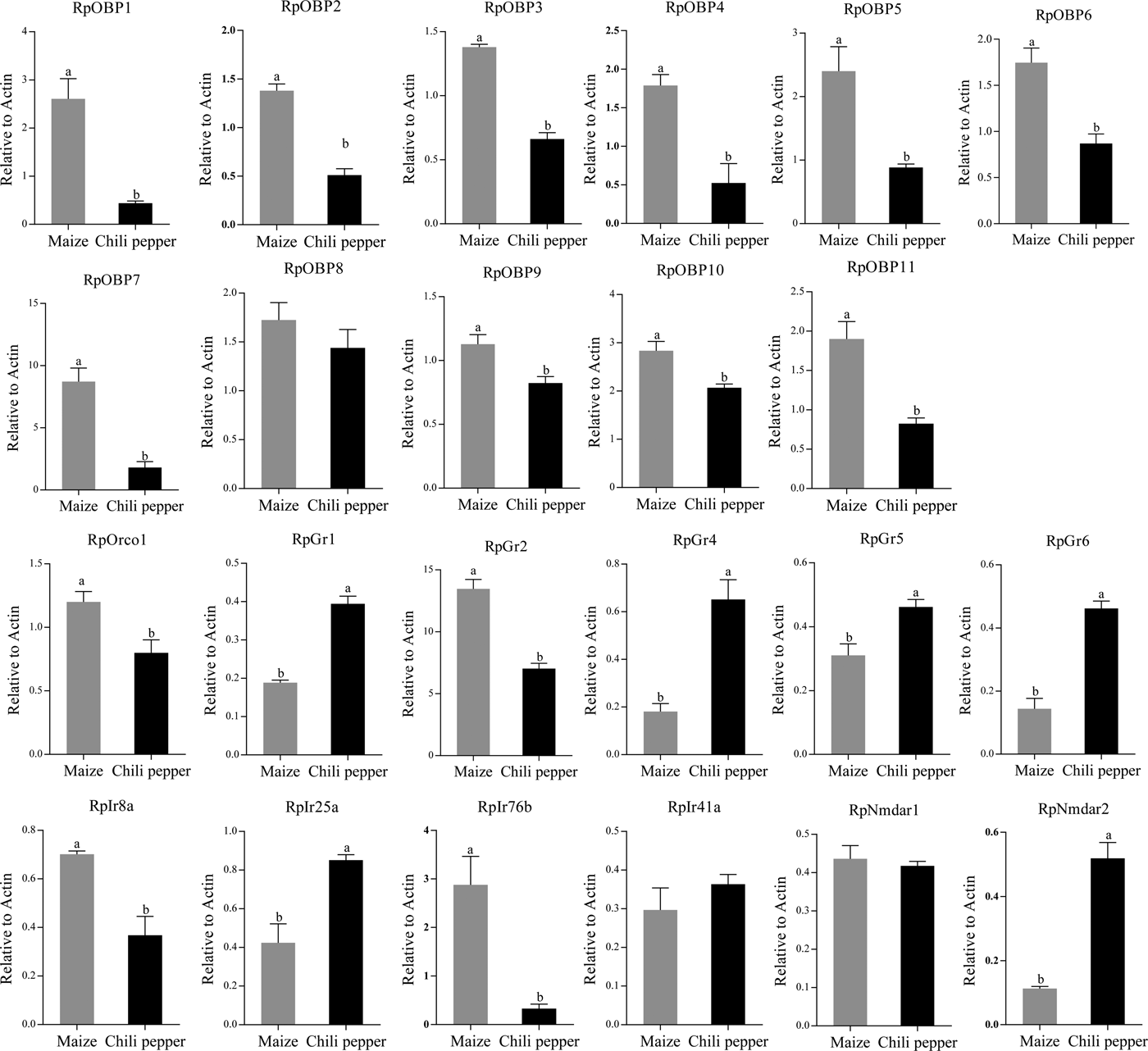
We verified the expression patterns of these chemosensory genes in different tissues, wing morphs and host plants. RpOBP1, RpOBP3 and RpOBP5 predominately expressed in body compared with head with antennae, while the rest selected genes, except RpOBP4, highly expressed in the head with antennae (fig. 5). When transferred to non-host plants (chili pepper), the expression of RpGr1, RpGr4, RpGr5, RpGr6, RpIr25a and RpNmdar2 in R. padi reared on maize were lower than that in R. padi reared on chili pepper (fig. 6). In the contrary, the expression of RpOBP1, RpOBP2, RpOBP3, RpOBP4, RpOBP5, RpOBP6, RpOBP7, RpOBP9, RpOBP10, RpOBP11, RpORco, RpGr2, RpIr8a and RpIr76b reared on maize were higher than that in R. padi reared on the chili pepper. RpOBP2, RpOBP5, RpOBP6, RpOBP7, RpOBP8 and RpOBP10 were abundantly expressed in winged aphid compared with wingless aphid whereas RpIr8a, RpIr25a, RpIr41a, RpIr76b, RpNmdar1 and RpNmdar2 predominately expressed in wingless aphid (fig. S3).
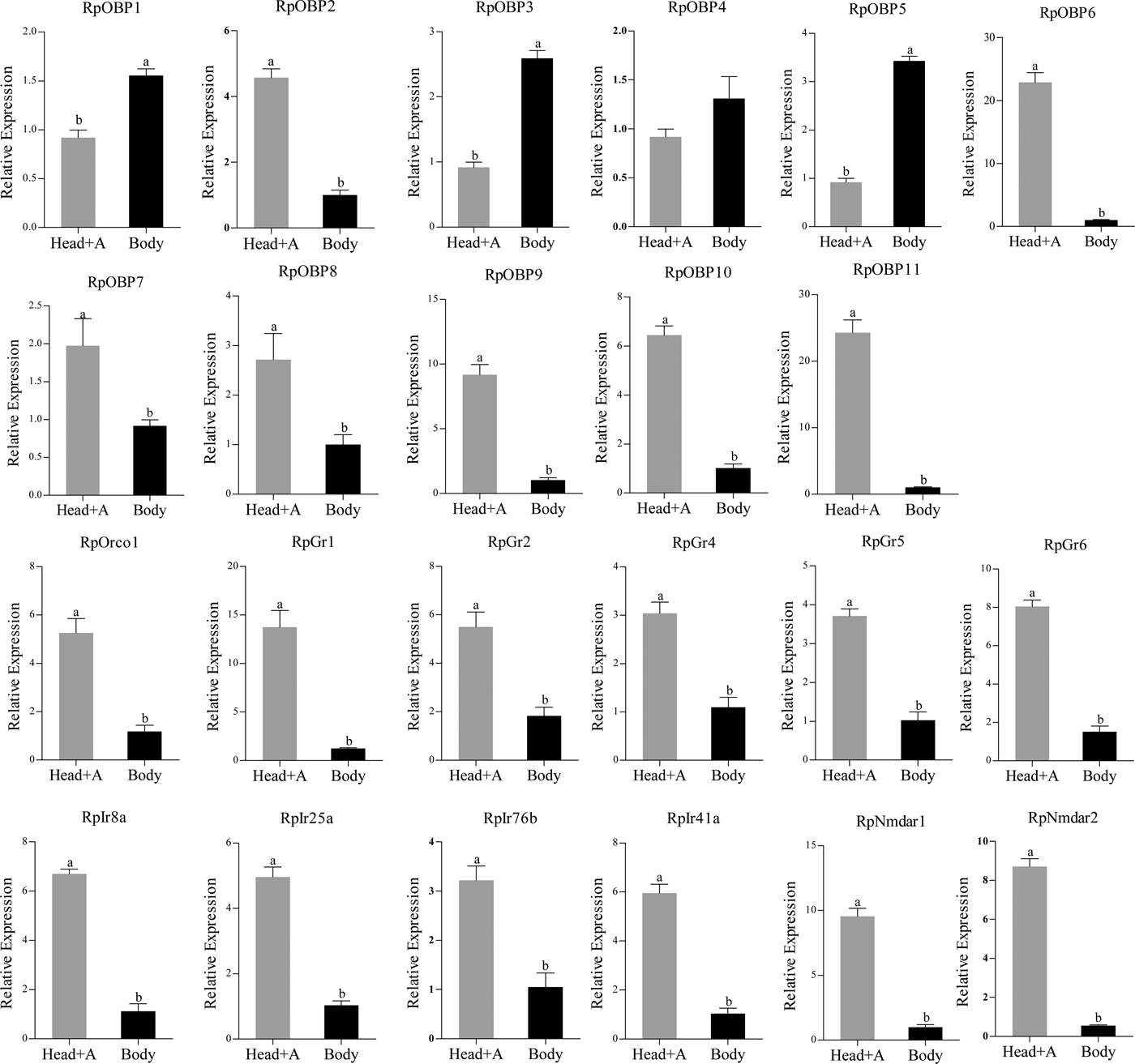
Fig. 5. The expression patterns of R. padi chemosensory genes in different tissues. Head+A: head with antennae; Body: only body without head and antennae. Data are presented as the mean of three replicates (n = 3) ± SE. The error bar represents standard error and the different small letters above each bar indicate significant differences in transcript abundances (P < 0.05).
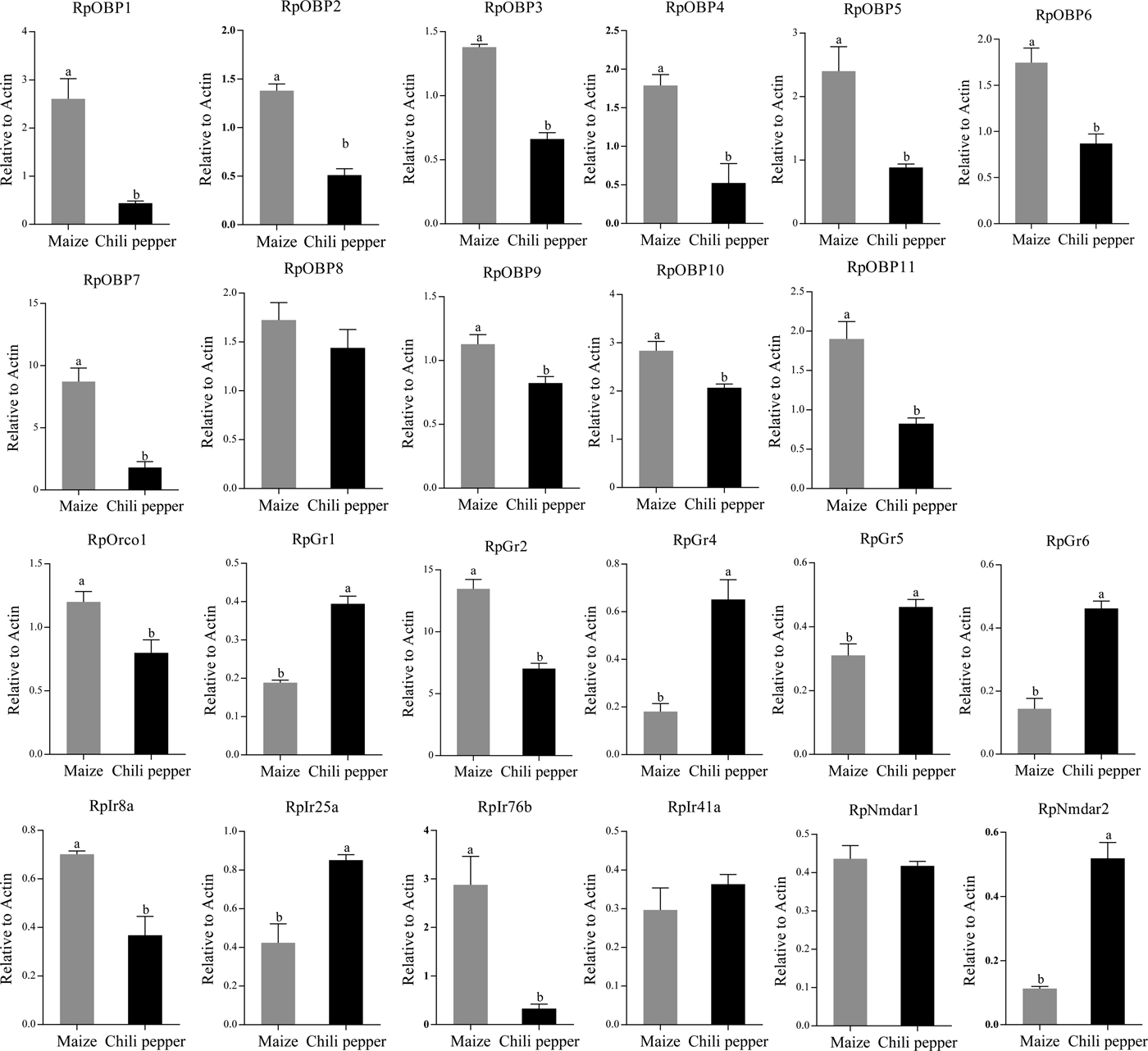
Fig. 6. The expression patterns of R. padi chemosensory genes in different host plants. Data are presented as the mean of three replicates (n = 3) ± SE. The error bar represents standard error and the different small letters above each bar indicate significant differences in transcript abundances (P < 0.05).
Discussion
Olfactory plays a critical role in insect survival and reproduction, such as foraging, aggregation, enemy avoidance, and mating (Hallem et al., Reference Hallem, Dahanukar and Carlson2006). Because of their season-dependent alternation of hosts, R. padi has gained massive attention for studying the host resistance and chemical cues in this process (Dixon, Reference Dixon1971). No investigation has focused on the putative functions of olfactory systems in this process. Our discoveries provide the first extensive molecular insights into the olfactory system of R. padi.
We performed a bioinformatic searching for olfactory genes based on the R. padi transcriptome using the related sequences from other species (Zhou et al., Reference Zhou, Vieira, He, Smadja, Liu, Rozas and Field2010; Cao et al., Reference Cao, Liu, Walker, Li and Wang2014; Xue et al., Reference Xue, Fan, Zhang, Xu, Han, Sun and Chen2016). Finally, we identified a total of 71 olfactory genes including 11 OBPs, nine CSPs, one SNMP, 15 ORs, 19 GRs and 16 IRs. All these candidates have been confirmed by Blastp in NCBI. The number of identified chemosensory genes in R. padi were similar with that in other aphid species except ORs, which was significantly less than that in other aphid species (Zhou et al., Reference Zhou, Vieira, He, Smadja, Liu, Rozas and Field2010; He et al., Reference He, Zhang, Liu, Zhang, Yang and Dong2011; Zhou et al., Reference Zhou, Min, Tang and Wang2015; Xue et al., Reference Xue, Fan, Zhang, Xu, Han, Sun and Chen2016). ORs identified in other aphid species were screened from the genome database. Thus, the less of identified ORs in R. padi might be the different sequencing strategy. And the sequencing tissues might be another reason for that (Kang et al., Reference Kang, Tian, Liu, Liu, Jing and Liu2017a). In this work, the transcriptome was conducted using the RNA extracted from the head with antennae.
As the first gate in the odorant recognition process, the functions of OBPs in various insect species have been well documented. For example, in the aphid, OBP3 and OBP7 in A. pisum and S. avenae were considered as the key delivery of aphid alarm pheromone whereas OBP1 only showed binding affinity with plant volatiles (Qiao et al., Reference Qiao, Tuccori, He, Gazzano, Field, Zhou and Pelosi2009). Furthermore, OBP1 and OBP3 were found to be expressed in cornicle (De Biasio et al., Reference De Biasio, Riviello, Bruno, Grimaldi, Congiu, Sun and Falabella2015). Consistent with this, in this work, we found RpOBP1 and RpOBP3 predominately expressed in body compared with head with antennae. And, RpOBP2, RpOBP5, RpOBP6, RpOBP7, RpOBP8 and RpOBP10 were abundantly expressed in winged aphid compared with wingless aphid. Similarly, when reared on the crowd condition to induce wing type aphid, the expression of OBP2, 6, 8 and 10 in A. pisum was significantly higher than that reared on the solitary condition (Vellichirammal et al., Reference Vellichirammal, Madayiputhiya and Brisson2016). Furthermore, when transferred to non-host plant, the expression of all the OBPs was significantly depressed except RpOBP8. As we all know, the plant quality, population density, alarm pheromones and interactions with predators, parasites will influence aphid to produce offspring with different wing morphs (winged or wingless) (Guo et al., Reference Guo, Zhang and Liu2016; Vellichirammal et al., Reference Vellichirammal, Madayiputhiya and Brisson2016). All of these results suggested that OBP might be essential in the wing morph determination signal reception process of aphid.
ORco was thought to be responsible for the OR adopting the correct structure and worked as a selective ion channel during olfactory signal transduction. In Dendroctonus armandi, the silencing of ORco led to EAG declining to 11 major volatiles of its host (Zhang et al., Reference Zhang, Gao and Chen2016a). RNAi and behavioral assay indicated that OR-based signaling pathway is mainly responsible for the attractive behavior of gregarious nymphs in the migratory locust (Wang et al., Reference Wang, Yang, Chen, Jiang, Li, Wang and Kang2015b). In S. avenae, (E)-beta-farnesene failed to induce production of winged aphids after the knockdown of SaOrco (Fan et al., Reference Fan, Zhang, Francis, Cheng, Sun and Chen2015). In our result, RpOrco1 abundantly expressed in the head with antennae and reared on maize significantly up-regulated its expression. However, there was no significant difference between the expression of RpOrco1 in the winged and wingless aphid. Besides the ORco1, there are 13 typical ORs in the R. padi transcriptome. And ApOr5 was identified as an essential agent to EBF reception in A. pisum (Zhang et al., Reference Zhang, Wang, Grossi, Falabella, Liu, Yan, Lu, Xi and Wang2016b). All of these results are consist with the previous work that ORs mediate the olfactory response to most food odors (Hallem et al., Reference Hallem, Dahanukar and Carlson2006).
As the different plant has different nutritional and defensive substances, and all of these substances were the last process for host-plant acceptance as a food source (Dewhirst & Pickett, Reference Dewhirst and Pickett2009; Nam & Hardie, Reference Nam and Hardie2014; Cao et al., Reference Cao, Liu, Zhang and Liu2016; Liu et al., Reference Liu, Kang, Guo, Zhu, Rahman Shah, Song, Fan, Jing and Liu2016). These aphid feeding behaviors are significantly regulated by these compounds. For instance, the feeding behaviors can be affected by sucrose, glycosides and glucosinolates (Takemura et al., Reference Takemura, Kuwahara and Nishida.2006; Kim & Jander, Reference Kim and Jander2007; Pescod et al., Reference Pescod, Quick and Douglas2007). And Grs are considered as the detector of these semiochemicals to co-ordinating insect feeding behaviors based on the life condition (Xu et al., Reference Xu, Papanicolaou, Zhang and Anderson2016). For example, B. mori, BmorGr6 was thought as a chemical sensor to influence the feeding behavior of B. mori larvae based on the food ingestion (Mang et al., Reference Mang, Shu, Endo, Yoshizawa, Nagata, Kikuta and Sato2016). In D. melanogaster, two Grs, Gr5a and Gr64a underlay the sugar perception result from the no physiological and behavioral response to any tested sugar in Gr5a and Gr64a mutant. And HaGr4 in H. armigera turned to D-fructose in antennal sensilla chaetica (Jiang et al., Reference Jiang, Ning, Guo, Jia, Huang, Qu and Wang2015). In this work, we found that the expression of RpGr1, RpGr4, RpGr5 and RpGr6 in R. padi reared on maize were lower than that of R. padi reared on the chili pepper. Contrary to this, RpGr2 predominant expressed in R. padi reared on maize. Furthermore, all of the selected Grs in R. padi predominately expressed in the head with antennae, which also revealed that these selected Grs were involved in the aphid gustatory reception. All of these results suggested that Gr might be crucial for the detection of the plant nutrition quality or defense substances.
Biogenic amines are necessary components of living cells, protecting cellular macromolecules and biomembrane phospholipids against damage under stress conditions. The accumulation of amines concentration cause the programmed cell death in response to various abiotic and biotic stresses such as herbivorous feeding. In previous work, plant amines participate in plant defence responses to R. padi through disturbing its feeding behavior by higher concentration (Sempruch et al., Reference Sempruch, Golawska, Osinski, Leszczynski, Czerniewicz, Sytykiewicz and Matok2016). And R. padi was able to reduce the accumulation of biogenic amines by depressed the activity of amino acid decarboxylation, which is a key step of biogenic amines biosynthesis (Sempruch et al., Reference Sempruch, Marczuk, Leszczyński and Czerniewicz2013). In addition, a previous work posited a model of that pea aphid can regulate the biogenic amine levels depending on their reared condition revealed by olfactory perception (Vellichirammal et al., Reference Vellichirammal, Madayiputhiya and Brisson2016). In insect olfaction systems, IR was considered to mediate responsiveness of OSNs to organic acids, amines and alcohol (Hussain et al., Reference Hussain, Zhang, Ucpunar, Svensson, Quillery, Gompel, Ignell and Grunwald Kadow2016). For example, AgIr76b mediated larval responses to butylamine (Liu et al., Reference Liu, Pitts, Bohbot, Jones, Wang and Zwiebel2010). In D. melanogaster, Ir20a mediates amino acid taste and blocks salt taste dependent on Ir76b (Ganguly et al., Reference Ganguly, Pang, Duong, Lee, Schoniger, Varady and Dahanukar2017). Furthermore, DmIr76b and DmGr66a were used to assess the quality and valence of polyamine in the supplied diet (Hussain et al., Reference Hussain, Zhang, Ucpunar, Svensson, Quillery, Gompel, Ignell and Grunwald Kadow2016). In this work, all of these selected IRs in R. padi were highly expressed in the head with antennae. Among them, the expression of RpIr8a and RpIr76b was lower in chili pepper reared aphid whereas the expression of RpIr25a and RpNmdar2 were higher in chili pepper reared aphid. And in Aphidius gifuensis, IRs also exhibited a host-specific expression patterns (Kang et al., Reference Kang, Tian, Liu, Liu, Jing and Liu2017a). All of these results indicated that IRs system in R. padi may be the detector of plant defense and involved in the regulation of host suitability along with GRs.
As the key role of the olfactory system in insect survival, it has been thought to be a target for pest management. For example, olfaction of RNAi treated aphids was severely damaged and the activity of these aphids were significantly restrained. And the injection of CqOr37/99-dsRNA in Culex quinquefasciatus significantly reduced the egg-laying induction of 4-ethylphenol (Zhu et al., Reference Zhu, Xu, Barbosa, Choo and Leal2013). Moreover, silenced RpOrco decreased blood-feeding volume, egg laying and molt rate in Rhodnius prolixus (Franco et al., Reference Franco, Oliveira, Moreira, Leal and Melo2016). In Apis cerana, neonicotinoid insecticides disrupted olfactory cognitive behavior (Li et al., Reference Li, Wu, Zhao, Tan, Jiang and Hu2015a; Tan et al., Reference Tan, Chen, Dong, Liu, Wang and Nieh2015). All of these results suggested that we could design the specific insecticides to disrupt the olfactory system in pest for controlling them. In addition, the key chemosensory receptor will be the target for designed or looking for the attractant, which might be used to kill pests combining with insecticides.
In this study, we not only identified the chemosensory genes in R. padi but also investigated the expression patterns of 23 selected genes among the different tissues, wing morphs and host plants. Combing with the demonstrated functions of related genes in other organisms, the tissue-, morph- and host-specific expression profile of these genes potentially revealed the candidate roles of these genes in the plant suitability assessment. Furthermore, the identification and expression patterns of chemosensory genes also provided initial background information for the further research on the molecular mechanism of the polyphagia and autumn migrants of it. Besides that, these chemosensory genes are also the candidate targets for pest management control in future.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485317001171
Author contributions
These studies were conceived of and designed by Z.W Kang and T.X Liu; Whole experiments were performed by Z.W Kang, F.H Liu, R.P Pang, W.B Yu and Z.Q Zheng; Data analysis and paper writing were done by Z.W Kang, F.H Liu, X.L Tan, H.G Tian and T.X Liu.
Conflict of interest disclosure
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the National Basic Research Program of Ministry of Science and Technology, China (973 Programs No. 2013CB127600), National Natural Science Foundation of China (No. 31272089 and 31471819), and China Agriculture Research System (CARS-25). The authors sincerely thank the James Hutton Institute for their submission of the HiSeq data and all staffs and students in the Key Laboratory of Applied Entomology, Northwest A&F University at Yangling, Shaanxi, China.