Introduction
Bluetongue (BT) and African horse sickness (AHS) are two non-contagious, arthropod-borne viral diseases that affect ruminants (both domestic and wild) and horses, respectively. Bluetongue disease is caused by the bluetongue virus (BTV), the prototype of the Orbivirus genus within the Reoviridae family, which also includes AHSV (Holmes et al., Reference Holmes, Boccardo, Estes, Furuichi, Murphy, Fauquet, Bishop, Ghabrial, Jarvis, Martelli, Mayo and Summers1995). Both viruses are transmitted by species of biting midges belonging to the Culicoides genus (Diptera: Ceratopogonidae) (Du Toit, Reference Du Toit1944; Mellor & Pitzolis, Reference Mellor and Pitzolis1979; Mellor et al., Reference Mellor, Boned, Hamblin and Graham1990) and are maintained naturally through a series of alternative cycles of replication between Culicoides vectors and susceptible hosts (Takamatsu et al., Reference Takamatsu, Mellor, Mertens, Kirkham, Burroughs and Parkhouse2003).
In the Mediterranean Basin and in southern Europe, the main vector for both viruses is Culicoides imicola (Mellor, Reference Mellor1996), although other Palaearctic Culicoides species, mainly within the subgenera Avaritia and Culicoides, contain midges (C. obsoletus, C. scoticus, C. dewulfi and C. pulicaris) known or suspected to be BTV vectors (De Liberato et al., Reference De Liberato, Scavia, Lorenzetti, Scaramozzino, Amaddeo, Cardeti, Scicluna, Ferrari and Autorino2005; Mehlhorn et al., Reference Mehlhorn, Walldorf, Klimpel, Jahn, Jaeger, Eschweiler, Hoffmann and Beer2007, Reference Mehlhorn, Walldorf, Klimpel, Schaub, Kiel, Focke, Liebisch, Liebisch, Werner, Bauer, Clausen, Bauer, Geier, Hörbrand, Bätza, Conraths, Hoffmann and Beer2009; Meiswinkel et al., Reference Meiswinkel, van Rijn, Leijs and Goffredo2007, Reference Meiswinkel, Baldet, de Deken, Takken, Delecolle and Mellor2008; Calvete et al., Reference Calvete, Calvo, Calavia, Miranda, Borrás, Estrada, Lucientes, Mañuz and Romero2008; Nielsen et al., Reference Nielsen, Nielsen and Chirico2010). In northern Europe and Spain, species of the Culicoides pulicares and the Culicoides obsoletus groups (C. obsoletus, C. scoticus and C. dewulfi) are involved in bluetongue pathology (Mehlhorn et al., Reference Mehlhorn, Walldorf, Klimpel, Schaub, Kiel, Focke, Liebisch, Liebisch, Werner, Bauer, Clausen, Bauer, Geier, Hörbrand, Bätza, Conraths, Hoffmann and Beer2009; Nielsen et al., Reference Nielsen, Nielsen and Chirico2010).
Blood meal identification provides information on host-feeding preferences or host-feeding patterns of insects in nature. Such information could indirectly provide data indicating which reservoirs are significant in vector-borne diseases (Lee et al., Reference Lee, Hassan, Hill, Cupp, Higazi, Mitchell, Godsey and Unnasch2002). Studies involving biting midges (Culicoides spp.) suggest that large mammals, specifically ruminants (Bartsch et al., Reference Bartsch, Bauer, Wiemann, Clausen and Steuber2009; Gerry et al., Reference Gerry, Sarto i Monteys, Moreno Vidal, Francino and Mullens2009; Mullens et al., Reference Mullens, Gerry, Monteys, Pinna and González2010), are the preferred host, but little is known about the transmission dynamics and the role of wild species. Bartsch et al. (Reference Bartsch, Bauer, Wiemann, Clausen and Steuber2009) determined by PCR that cattle, on selected farms with several species, were the most attractive host for Culicoides obsoletus and Pulicaris groups even if other large mammals were located in their immediate vicinity. The aim of the present study was to identify blood meals and establish feeding patterns of Culicoides spp. in Picos de Europa National Park in northern Spain, where livestock and wildlife coexist. Blood meals were identified by sequencing PCR products of the cytochrome b gene from mitochondrial DNA in Culicoides species.
Material and methods
Study area
The field work was conducted in the Picos de Europa National Park, 20km from the northern coast of Spain (centred on 43°15′N, 5°00′W) (fig. 1). The Picos de Europa is a predominantly limestone mountain range with a marked elevation gradient from 200 to 2600m a.s.l. The climate is extremely wet, particularly in the northern slopes, with precipitation values exceeding 1500mm year–1 in most sites. In the Picos de Europa National Park, extensive livestock rearing has been traditionally practised by small family-based units with an average of 10–12 cows and 50 sheep and/or goats (De Sebastian, Reference De Sebastián, Palacios and Llaneza1997). Transhumance is common, with animals moving seasonally from the valley bottoms, where they spend the coldest months of winter, to common mountain pastures in summer, where the livestock are allowed to free-range. Wildlife and livestock coexist in the study area.

Fig. 1. Location of the study area (northern Spain) and the geographical distribution (black dots) of sampled points. The digital elevation model of the area is shown in grey. Darker and clearer areas represent lower and higher altitudes, respectively.
Insect samples
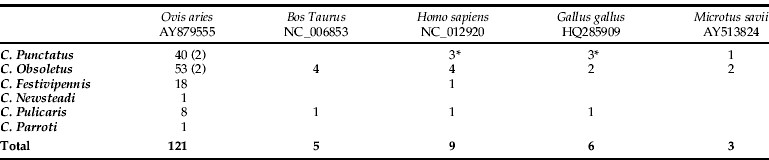
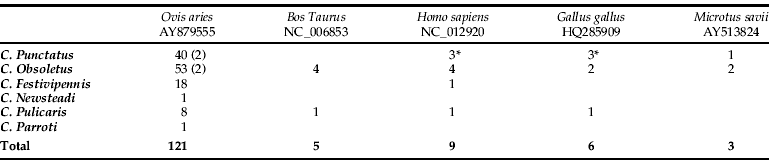
Trapping was performed using a 4 W ultraviolet light trap fitted with a suction fan, which is recommended to trap Culicoides spp. (Miniature Blacklight Model 1212, John Hock Company, Gainesville, FL, USA). Three traps were placed 1.7 to 2.5m above ground level near riverbanks and pastures (fig. 1). Trap 1 was located at 1057ma.s.l. and close to a shepherd house which is occupied in summer. Traps 2 and 3 were located at 842 and 1062ma.s.l., respectively. All collected insects were transferred into micro tubes, preserved in 70% ethanol and then transported to the laboratory. Tubes were kept frozen (−20 °C) until species identification and DNA extraction. In our work, time between capturing and processing the blood meal analysis were four months. Upon examination, species of Culicoides were separated from other insects, with identification to species level based on previously described wing patterns (Rawlings, Reference Rawlings1996). In the summer of 2008, 158 fully fed females were randomly selected (table 1) for further analysis. In this paper, the Culicoides obsoletus group refers to three putative species: C. obsoletus, C. scoticus and C. dewulfi.
Table 1. Blood meal sources from Culicoides species. GenBank accession numbers are indicated.
Asterisk shows one midge that has acquired blood meals from humans and chickens. Numbers in brackets indicate number of midges with blood from two different sheep.
DNA extraction, polymerase chain reaction (PCR) amplification and DNA sequencing
Total DNA was extracted from single midges using a commercial kit (Nucleo Spin Tissue, Mancherey-Nagel, Düren, Germany). Isolated DNA from the midge and blood-meals served as a template for subsequent PCR amplification. All DNA templates were screened with avian and mammalian specific primer pairs for the cytochrome b gene (cytB) with expected product sizes of 508 and 772bp, respectively (Molaei et al., Reference Molaei, Oliver, Andreadis, Armstrong and Howard2006). Primers used for specific avian amplification were 5′-GAC TGT GAC AAA ATC CCN TTC CA-3′(forward) and 5′-GGT CTT CAT CTY HGG YTT ACAAGA C-3′(reverse). Mammalian specific PCR primers were 5′-CGA AGC TTG ATA TGA AAA ACC ATC GTT G-3′(forward) and 5′-TGT AGT TRT CWG GGT CHC CTA-3′(reverse). Because of sequence homology among classes, these primer sets also amplify reptilian DNA. PCR was performed in a final volume of 25μl containing 200ng of total DNA, 10pmol of each primer, 200μM dNTPs, 2.5mM MgCl2, 50mM KCl, 10mM Tris-HCl, 0.1% Triton X-100, 0.1μl of PCR enhancer (wondertool, Biotools, Spain) and 1U Taq + 0.5U Pfupolymerases (Biotools, Spain). Forty-five cycles were performed with the following step-cycle program: strand denaturation at 94 °C for 45s, primer annealing at 60 °C for avian and 57 °C, for mammalian primer pairs for 45s, and primer extension at 72 °C for 40s. Two PCR amplifications were carried out for avian or mammalian discrimination in each specimen, obtaining a product size of 508 and 772bp, respectively. Positive PCR products were purified using a commercial kit (Nucleo Spin Extract, Mancherey-Nagel, Düren, Germany) and were sequenced using an ABI Prism 3700 (Applied Biosystems). PCR products were directly sequenced using the primers described above. All amplified products were sequenced from both strands. Sequences were identified using BLASTn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches in the GenBank database to compare fragments. Only matches with at least 98% were accepted.
Results and discussion
We identify the host species by DNA sequencing in 144 of 170 (84.7%) specimens tested (table 1). In the remaining samples, identification of the blood-meal source was unsuccessful, possibly due to the post-ingestion time prior to sampling, time between capturing and processing the blood meal analysis or availability of the species-specific cytochrome b gene sequences in the database. In our study, eight of the 23 unsuccessful attempts at identifying the blood-meal source used mammalian specific primer pairs. The amplified product generated weak bands, and it was not possible to obtain high quality sequences to compare fragments in GenBank, suggesting that host DNA concentrations were low or degraded in the source extractions. Additionally, several studies have demonstrated the important effects that post-ingestion time and the physical conditions under which sacrificed insects are stored can have on the success of detecting blood meal DNA (Mukabana et al., Reference Mukabana, Takken, Seda, Killeen, Hawley and Knols2002; Oshaghi et al., Reference Oshaghi, Chavshin, Vatandoost, Yaaghoobi, Mohtarami and Noorjah2006; Haouas et al., Reference Haouas, Pesson, Boudabous, Dedet, Babba and Ravel2007). Oshaghi et al. (Reference Oshaghi, Chavshin, Vatandoost, Yaaghoobi, Mohtarami and Noorjah2006) showed that host DNA extracted from a blood meal up to 33h post-ingestion acts as an efficient template for PCR amplification. However, meals digested for 36h or longer required higher DNA concentrations to obtain successful PCR amplification. Haouas et al. (Reference Haouas, Pesson, Boudabous, Dedet, Babba and Ravel2007) found a DNA detection sensitivity varying from 100% to 66% for blood digested from 0–18h to 24–72h, respectively, with an overall of 79% positive results in the blood-meal PCR assay. In our study, overall, 84.7% of the blood fed midges showed positive identification. It is also highlighted that the sensitivity depends on both the quantity and degradation of target DNA. Culicoides spp. vary in size within and between species, and blood meal volume varies from 0.01 to 0.06μl (Venter et al., Reference Venter, Paweska, Lunt, Mellor and Carpenter2005). This blood meal volume is lower than many other insects, such as sand flies (1μl) (Haouas et al., Reference Haouas, Pesson, Boudabous, Dedet, Babba and Ravel2007), tsetse flies (37μl) (Torr & Hargrove, Reference Torr and Hargrove1998) or anopheline mosquitoes, which are able to concentrate erythrocytes during the blood feeding process (Briegel & Rezzonico, Reference Briegel and Rezzonico1985).
Furthermore, only matches with at least 98% were accepted in our study; three samples did not match exactly the DNA sequences available in the GenBank database. Specifically, sequence matches with 93%, 95% and 91% identity with Lamprolepis smaragdina (GenBank accession number AY217803) (C. obsoletus), Tragulus javanicus (GenBank accession number AY121994) (C. pulicaris) and Francolinus hartlaubi (GenBank accession number U90639) (C. festivipennis), respectively, were found. Because these species of wildlife are not known to be present in Spain, the results indicate that the blood host identity for these midges could be a related species, but we could not conclude which one.
The majority of identified blood meals were derived from mammalian blood (95.8%), and only six contained chicken blood. These blood meals may have come from chickens (Gallus gallus; HQ285909) that are part of a conservation programme of an autochthonous chicken breed that is settled in the Picos de Europa National Park. We identify five species as mammalian hosts for Culicoides spp. A majority of the blood meals (n = 121, 87.7%) were from sheep, followed by human (Homo sapiens; GenBank accession number NC_012920) (n = 9, 6.5%), cattle (Bos taurus; GenBank accession number NC_006853) (n = 5, 3.7%), and Savi's Pine Vole (Micrototus savii; GenBank accession number AY513824) (n = 3, 2.1%). Table 1 shows the number of blood meals identified from each of the Culicoides spp. sampled in the Picos de Europa National Park in the summer of 2008. This study found that large mammals, specifically ruminants, were the preferred host for biting midges (Culicoides spp.), but opportunistic feeding behaviour was also found (human, chicken and Savi's Pine Vole were also identified). Host feeding behaviour of Culicoides species may also be influenced by the relative abundance of other possible host species in the study area. In this regard, the wild species, Savi's Pine Vole (Micrototus savii), was found to be of relevance as a host and as a putative reservoir for viruses transmitted by species of biting midges belonging to the Culicoides genus. Furthermore, the DNA sequencing results from the three blood-meal species showed high identity for three wild species (Lamprolepis smaragdina, Tragalus javanicus and Francolinus hartlaubi), providing additional support for the existence of opportunistic feeding behaviour on non-domestic species. Bartsch et al. (Reference Bartsch, Bauer, Wiemann, Clausen and Steuber2009) determined that cattle located on farms with multiple species were the most attractive host for C. obsoletus and Pulicaris groups, even if other large mammals were located in the immediate vicinity. Surprisingly, none of the biting midges sampled in that study were found to be positive for sheep's blood. This finding differs from our work and is most likely reflective of the blood meal source being dependent on the attractiveness and the numeric availability of the host. In the Picos de Europa National Park, domestic sheep are much more numerous than cattle. Midges fed on humans and chicken were trapped in the same sample point, close to a shepherd house which is occupied in summer. This was the place where chicken was located.
Other described methods, such as PCR-RFLPs (Maleki-Ravasan et al., Reference Maleki-Ravasan, Oshaghi, Javadian, Rassi, Sadraei and Mohtarami2009; Oshaghi et al., Reference Oshaghi, Maleki Ravasan, Hide, Javadian, Rassi, Sadraei, Mohebali, Sedaghat, Hajjaran, Zarei and Mohtarami2009) or multiplex PCR (Haouas et al., Reference Haouas, Pesson, Boudabous, Dedet, Babba and Ravel2007) are based on identification of inter specific polymorphisms in the cytochrome b gene between species, and the digestion with specific restriction enzymes or the design of specific primers, respectively. In this works, the blood meal origin was identified obtaining different specific species profiles. However, the RLFP or multiplex PCR techniques are based on multiple alignments of the mitochondrial cytochrome b gene, and then only species from the multiple alignment can be identify. In our work, the sequencing method used spends more time in the identification but has the advantage to be able to detect any species with an identified sequence deposited in GenBank, including wild species.
Finally, the results showed that a surprising 3.4% of blood-meals were from two different hosts. One midge may have acquired blood meals from human and chicken sources, and then specifics bands of 508 and 772bp for avian and mammalian amplifications were obtained, respectively. Sequence data confirmed the two different origin of the blood. Four other midges fed on two different sheep, as evidenced by the observation of two distinct sheep haplotypes (fig. 2), and confirmed by sequencing of the PCR products by the forward and reverse primers. The cytochrome b gene is located in the mitochondrial DNA, which is haploid and maternally inherited, and heterozygous mitochondrial DNA is very rare and not reported in sheep. However, several studies have demonstrated the presence of leaked paternal mitochondrial DNA (biparental transmission of mitochondrial DNA), particularly in interspecific hybrids between closely related species in several genera, such as Mytilus (Zouros et al., Reference Zouros, Oberhauser Ball, Saavedra and Freeman1994; Rawson et al., Reference Rawson, Secor and Hilbish1996), Drosophila (Kondo et al., Reference Kondo, Satta, Matsuura, Ishiwa, Takahata and Chigusa1990) and Mus (Gyllensten et al., Reference Gyllensten, Wharton, Josefsson and Wilson1991; Kaneda et al., Reference Kaneda, Hayashi, Takahama, Taya, Lindahl and Yonekawa1995). The observation of a heterozygous nucleotide in the sequencing chromatogram could indicate two haplotypes and then multiple feeding. Homology searches of the two haplotypes showed 100% identity with sheep. Furthermore, this substitution was conservative, and no amino acid change was observed in the protein sequence between the two haplotypes. Multiple feedings on potential hosts within the same gonotrophic cycle may enhance amplification and transmission of viruses carried by these vectors. However, multiple feeding may or may not favour disease transmission, depending on whether the vector feeds on potential or dead-end hosts (Arunachalam et al., Reference Arunachalam, Samuel, Hiriyan, Rajendran and Dash2005).

Fig. 2. Sequencing chromatograms obtained by sequencing of the PCR products by the (a) forward and (b) reverse primers of a midge fed on two different sheep. Arrow shows heterozygous position, and then two different haplotypes.
In conclusion, these results reveal that biting midges of Culicoides spp. can feed on a variety of vertebrate hosts including mammals, birds and wild species, with a preference for ruminants. Evidence for opportunistic feeding behaviour on wild species was found along with feeding on multiple potential hosts.
Acknowledgements
The study was supported by the Picos de Europa National Park Funds. The authors wish to thank Miguel Menández de la Hoz and the Picos de Europa National Park staff for their technical assistance and for providing the experimental samples.