Introduction
Aphids cause serious economic damage to cereal crops by transmitting viruses and, in outbreak years, by reducing yields through direct feeding (van Emden & Harrington, Reference van Emden and Harrington2007). Fortunately, cereal aphids are attacked by a suite of natural enemies, including parasitoids and generalist predators (Chambers et al., Reference Chambers, Sunderland, Stacey and Wyatt1986; Sunderland et al., Reference Sunderland, Axelsen, Dromph, Freier, Hemptinne, Holst, Mols, Petersen, Powell, Ruggle, Triltsch and Winder1997), both of which have been shown to be capable of reducing aphid population densities (Schmidt et al., Reference Schmidt, Lauer, Purtauf, Thies, Schaefer and Tscharntke2003, Reference Schmidt, Thewes, Thies and Tscharntke2004; Bell et al., Reference Bell, Traugott, Sunderland, Skirvin, Mead, Kravar-Garde, Reynolds, Fenlon and Symondson2008a; Macfadyen et al., Reference Macfadyen, Gibson, Polaszek, Morris, Craze, Planque, Symondson and Memmott2009). This ecosystem service provides economic benefits to farmers. For example, the yield increase in spring barley attributable to generalist predators was found to be comparable with the yield increases from insecticide use (Östman et al., Reference Östman, Ekbom and Bengtsson2003). However, generalist predators do not only feed on the pest, but might also consume parasitoids, attenuating the overall pest suppression effect (Brodeur & Rosenheim, Reference Brodeur and Rosenheim2000; Snyder & Ives, Reference Snyder and Ives2001). Empirical evidence of this type of intraguild predation (IGP) stems either from studies employing observational approaches or cage experiments, where the assessment of feeding interactions is limited to particular taxa and where trophic connections are indirectly inferred. Although empirically established aphid-parasitoid food webs have been described previously (e.g. Müller et al., Reference Müller, Adriaanse, Belshaw and Godfray1999; van Veen et al., Reference van Veen, Müller, Pell and Godfray2008), polyphagous predators have yet to be integrated in these trophic networks to examine how they are connected to aphids and their parasitoids (but see Bell et al., Reference Bell, King, Bohan and Symondson2010). This is mainly due to the practical difficulties inherent in identifying and quantifying predator-prey interactions from field data, especially for highly polyphagous species with less predictable diets (Symondson, Reference Symondson2002; van Veen et al., Reference van Veen, Morris and Godfray2006). Their exclusion, however, considerably limits our understanding of the functioning of these natural enemy communities where generalist predators may regularly interfere with parasitoid aphid control. If generalist predators consume parasitoids and their hosts at random, this would have little effect on aphid-parasitoid interactions. However, random feeding by generalists is highly unlikely (Agusti et al., Reference Agustí, Shayler, Harwood, Vaughan, Sunderland and Symondson2003; Harwood et al., Reference Harwood, Desneux, Yoo, Rowley, Greenstone, Obrycki and O'Neil2007; Juen & Traugott, Reference Juen and Traugott2007; Kuusk et al., Reference Kuusk, Cassel-Lundhagen, Kvarnheden and Ekbom2008; King et al., Reference King, Vaughan, Bell, Bohan and Symondson2010; Eitzinger & Traugott, Reference Eitzinger and Traugott2011). If, instead, generalist predators demonstrate prey choice, aphid-parasitoid trophic interactions might be altered in ways that are not affected by internal aphid-parasitoid dynamics.
The aim of this study was to examine the trophic structuring within an aphid-parasitoid-generalist predator community using a molecular approach and to measure how frequently parasitoid and aphid prey are consumed by generalist predators such as carabid and staphylinid beetles or spiders. Our objectives were, therefore, three-fold: (i) to use PCR (polymerase chain reaction) to identify and measure the frequency of trophic interactions between aphids, parasitoids and predators, (ii) to examine how predation on aphids and parasitoids changes during aphid population growth, and (iii) to assess which predators are most likely to interfere with aphid control by parasitoids.
Materials and methods
Experimental site and field sampling
Field work was conducted in a 13-ha winter wheat field at the Warwick HRI experimental farm, Wellesbourne, Warwickshire, UK (52°12.18′N, 1°36.00′W) within a larger experiment investigating the effects of compost addition on invertebrate predators and their prey. For a detailed description of how communities were sampled, see Bell et al. (Reference Bell, Traugott, Sunderland, Skirvin, Mead, Kravar-Garde, Reynolds, Fenlon and Symondson2008a). Here, we focus on the aphid-parasitoid-predator food webs and combine the data from different compost treatments.
Ground-active predators were collected within quadrats (0.5×0.5 m) using aspirators. Predators were taken from two quadrats within 16 experimental plots, totalling 32 quadrat samples per date. Quadrat sampling began on 24th May and ended on 3rd July 2005 (four collection dates), covering the period of the build-up of the aphid population. Predators were placed individually in 1.5-ml reaction tubes, put on ice and frozen within 3 h at −80°C.
Aphid population densities were estimated over the 12-week period from 2nd May to 18th July. Total numbers of aphids were counted from four randomly selected wheat tillers per plot. Sitobion avenae (F.) represented the great majority of aphids, and other cereal aphid species, such as Rhopalosiphum padi (L.) and Metopolophium dirhodum (Walker), occurred at very low densities. All aphids collected were individually frozen at –80°C and all S. avenae screened for parasitoid DNA to determine the rates of parasitism by each parasitoid species on each sampling date (Traugott et al., Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008).
In addition to the sampling program described above, arthropods occurring in the wheat field were also collected for cross-amplification testing with the aphid- and parasitoid-specific primers, using hand sampling, micro-aspirators or sweep nets. All non-predatory arthropods collected were freeze-killed on capture and stored in 80% ethanol for subsequent DNA extraction. Predators were handled differently to avoid the contaminating effects of parasitoid or aphid remains amongst their gut contents. These were starved for one week at room temperature to allow digestion and clearance of their guts, freeze-killed and then stored in 80% ethanol. Thereafter, these arthropod samples were DNA-extracted and used for cross-amplification testing, using the PCR assays described below.
DNA extraction
The DNA of parasitoids that emerged from aphids (Traugott et al., Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008), and other field-collected invertebrates used for cross-amplification testing, was extracted using a modified Chelex extraction protocol. The DNA of field-collected predators (whole animals) was extracted using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions; 200 μl DNA extracts were stored at –24°C. Two extraction-negative controls were included in every batch of 48 samples and tested for DNA carry-over contamination using universal invertebrate primers as described in Traugott et al. (Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008).
Diagnostic multiplex and singleplex PCR
A multiplex-PCR system was developed, allowing us to simultaneously screen predator gut contents for parasitoid and aphid DNA. Primer pairs, targeting S. avenae (S103/A103), the aphid parasitoids Ephedrus plagiator (Nees) (S115/A115) and four species of Aphidius (S108/A107), as well as the aphid hyperparasitoid Dendrocerus carpenteri (Curtis) (S120/A122), were designed from cytochrome c oxidase subunit I gene (COI) sequences (see table S1 in the supplementary material). Primers designed for use in the multiplex-PCR amplified short fragments, from 114 bp to 231 bp, in order to increase prey DNA detection success (King et al., Reference King, Read, Traugott and Symondson2008). Optimisation of the PCR protocols included determination of optimum annealing temperatures by temperature-gradient PCR, testing different concentrations and combinations of primers in multiplex PCR and adjusting cycling conditions. Each 10 μl multiplex-PCR reaction mix contained 2.5 μl of extracted DNA, 1×multiplex-PCR master mix (Qiagen), 1.5 μl of PCR-water (Qiagen) and the primers at their respective concentration (table S1). The cycling conditions were 15 min at 95°C, 35 cycles of 30 s at 94°C, 3 min at 62.5°C, 1 min at 72°C and final elongation for 10 min at 72°C.
As the multiplex PCR included a group-specific primer pair, covering four Aphidius species detected in the field-collected aphids (Traugott et al., Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008), all aphids testing positive with this primer pair were retested in singleplex PCRs with species-specific primers to identify the respective Aphidius species (table S1). The six parasitoid species targeted in both singleplex- and multiplex-PCR assays accounted for 84.3% of all parasitoids detected by diagnostic PCR within the aphids sampled (Traugott et al., Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008), thus enabling us to measure predation on the most abundant parasitoid species. To confirm the results obtained by multiplex PCR, each sample testing positive for parasitoid or aphid DNA was re-tested in singleplex PCR using the appropriate primer pair. To ensure comparability between multiplex- and singleplex-PCR assays, the cycling conditions and reaction mix, as described above for multiplex PCR, were used, except that primer concentration was 1 μM. Within all PCRs, 1:10 diluted DNA of the target species was included as a positive control along with a minimum of four negative controls (DNA-free H2O). All predator DNA extracts which tested negative for any of the prey species described above were tested for their ability to amplify in PCR as described in Traugott et al. (Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008); 3.0% of DNA extracts did not amplify in these tests and were excluded from the analysis. All PCR assays were carried out in Eppendorf (Hamburg, Germany) Mastercyclers.
PCR assay specificity and sensitivity
Diagnostic analyses of both predation and parasitism require extensive cross-amplification tests to ensure their specificity (King et al., Reference King, Read, Traugott and Symondson2008). These tests were conducted for both multiplex- and singleplex-PCR assays and included S. avenae and the six aphid parasitoid species tested for, as well as five additional aphid species, four additional aphid and 15 non-aphid parasitoids, and other plant- and soil-dwelling invertebrates collected within the experimental field (see table S2). Before conducting the cross-amplification tests, DNA extracts from all taxa listed within tables S1 and S2 were tested for their ability to amplify in PCR using universal invertebrate primers (see above).
Primer sensitivity was determined for all primer pairs used in the singleplex and multiplex PCRs by testing dilutions of target DNA for amplification success (for the group-specific Aphidius primer pair DNA from Aphidius rhopalosiphi De Stefani was used). DNA concentration in the original extracts was determined using NanoDrop (NanoDrop Technologies, Wilmington, DE, USA), adjusted to 0.5 ng μl−1 and two-fold serially diluted. The serial diluted target DNA was then used as template in the multiplex and singleplex assays at concentrations of 62.5, 31.3, 15.6, 7.8, 3.9, 1.9, 0.98, 0.49, 0.24, 0.12, 0.061, 0.031 and 0.015 pg of parasitoid DNA per μl PCR.
PCR products were separated and detected using the QIAxcel System and QIAxcel DNA screening kit (Qiagen) with separation method AL320. Electropherograms were analysed and scored using BioCalculator Fast Analysis Software version 3.0 (Qiagen); all samples generating >0.1 fluorescent units (which is well above the cartridges' background fluorescence-induced error) were deemed to be positive. The fragment lengths of PCR products amplified from field-collected predators were determined by comparing them with PCR-fragments from the positive controls to reliably score amplified aphid and parasitoid DNA.
Analysis
Predator densities, as established from quadrat sampling, were compared between sampling dates within each predator group and for the pooled predator catches using Kruskal-Wallis ANOVA and Mann-Whitney U tests for pair-wise comparisons.
Parasitism rates for S. avenae are based on a previous analysis of aphid parasitism in the same winter wheat field (Traugott et al., Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008) using the data for the six parasitoid species targeted in the present paper. For prey detection rates in predators and parasitoid detection rates in S. avenae, 95% tilting confidence limits (CL), which adjust for bias and skewness in the bootstrap distribution and are asymmetrical (Hesterberg et al., Reference Hesterberg, Moore, Monaghan, Clipson, Epstein, Moore and McCabe2003), were calculated by 9999 bootstrap resamples using S-PLUS 8.0 (Insightful Corporations, Seattle, WA, USA). To account for the significantly longer prey DNA detection times regularly found in spiders compared to carabid beetles (Sheppard et al., Reference Sheppard, Bell, Sunderland, Fenlon, Skervin and Symondson2005; Traugott & Symondson, Reference Traugott and Symondson2008; Sint et al., Reference Sint, Raso, Kaufmann and Traugott2011), prey detection rates were down-weighted in spiders to allow for more accurate comparison with prey detection in beetles. Down-weighting was based on the feeding experiments by Traugott & Symondson (Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008), in which aphid and parasitoid DNA detection success was found to be on average 48.2% (i.e. a factor of 2.1) higher in Erigone sp. compared to the carabid Demetrias atricapillus. This calculation is based on the differences in prey DNA detection between spiders and beetles observed for the time points 0, 8, 16, 24 and 32 h post-feeding for a 369 bp aphid fragment and a 291 bp parasitoid fragment. The difference in amplification success was calculated for each time point; and then the mean was calculated for the differences observed, leading to the 48.2% enhanced detection success in spiders as reported above. Hence, prey DNA detection in spiders was divided by 2.1, and 95% tilting confidence limits were recalculated. Data from other feeding experiments where Tenuiphantes tenuis and Tachyporus sp. were fed with parasitized aphids (M. Traugott & W.O.C. Symondson, unpublished data) suggest a very similar relationship.
IGP on parasitoids can either be direct, when predators capture and eat adult parasitoids, or coincidental, when parasitized aphids are eaten (Polis et al., Reference Polis, Myers and Holt1989). Unfortunately, current DNA-based methodology does not allow us to differentiate between these two scenarios: predators which test positive for host and parasitoid DNA could have eaten a parasitized host or the prey DNA might stem from separate host and parasitoid meals. We considered predators which tested positive for parasitoid DNA only to be a conservative proxy for direct IGP, as aphid DNA is usually detectable as well when parasitized and mummified aphids are eaten (Traugott & Symondson, Reference Traugott and Symondson2008; Traugott et al., Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008). To estimate predation on unparasitized aphids, predators which tested positive for aphid DNA only were included, providing a proxy for aphid mortality due to predation. For both, the estimation of direct IGP and predation on unparasitized aphids, all four sampling dates were pooled. Note, however, that the estimated predation rates on unparasitized aphids might be overestimated due to consumption of aphids containing very early instar parasitoids, which are hard to detect using PCR-based approaches (Traugott & Symondson, Reference Traugott and Symondson2008). On the other hand, predation on unparasitized aphids and on adult parasitoids might be underestimated due to predators which consumed aphids and parasitoids in separate meals, which corresponds in our analysis to the consumption of parasitized aphids.
Results
The invertebrate predator community
The invertebrate predators collected within the quadrats (1255 individuals from 61 taxa; for details of species see table S3) were dominated by carabid beetles (53%), spiders (30%) and staphylinid beetles (12%). Mean predator densities were similar on 24 May and 21 June (∼5 individuals 0.25 m−2), whereas they doubled and tripled on 8 June and 3 July, respectively (fig. 1). Among the carabids, Trechus quadristriatus Schrank and Notiophilus biguttatus (F.) were most abundant and accounted for 59% and 24% of all ground beetles caught, respectively. Of the spiders caught, 95% belonged to the Linyphiidae. Apart from unidentified juveniles (45%), three species dominated within the linyphiids: Erigone atra (Blackwall) (17%), Tenuiphantes tenuis (Blackwall) (14%) and Bathyphantes gracilis (Blackwall) (10%). Tachyporus hypnorum (F.) (29%) was the most frequently collected staphylinid beetle. As most of the beetle and spider species were found only in low numbers, too few individuals were available for a meaningful predator-specific dietary analysis. Hence, the predators were amalgamated into five groups based on their abundance and functional attributes (Bell et al., Reference Bell, Mead, Skirvin, Sunderland, Fenlon and Symondson2008b): (i) T. quadristriatus, (ii) N. biguttatus, (iii) Coccinellidae, Cantharidae and other Carabidae (subsequently referred to as ‘other beetles’), (iv) Staphylinidae and (v) spiders. Within each group, the magnitude of the average rank densities changed significantly between the sampling dates, with most pronounced differences in T. quadristriatus (fig. 1).

Fig. 1. Mean densities of arthropod predators sampled at four dates between 24 May and 3 July 2005 within a winter wheat field near Warwick, UK. At each date, mean arthropod densities were established from 36 quadrat (0.5×0.5 m) samples. Different letters denote significant differences in predator densities at P<0.05.
PCR specificity and sensitivity
Both the singleplex- and multiplex-PCR assays proved to be highly specific as no cross-amplification was found when the non-target species were tested. The sensitivities of the different primers used within the multiplex PCR differed only marginally (see appendix S1 in the supplementary material).
Predation on aphids
Numbers of S. avenae increased monotonically with sampling date and peaked in July at a mean density of 25.9 (±5.2 SE) aphids per four wheat tillers (fig. 2a). Although no aphids were detected on the wheat plants sampled on 24 May, 28.0% of all predators tested positive for DNA of S. avenae (fig. 2a, note that detection rates in spiders were down-weighted), showing that the predators were preying on aphids even at low densities during their establishment phase. Aphid detection rates peaked on 21 June, when 33.3% of predators tested positive (fig. 2a). Overall, the highest proportion of predators testing positive for S. avenae was found in N. biguttatus (64.0%), whereas less positives were obtained in other beetles (50.4%) and staphylinids (38.0%). Compared to N. biguttatus, a significant smaller proportion of spiders (18.9%, down-weighted) and T. quadristriatus (5.8%) contained DNA of S. avenae (table 1). Although there were significant differences in aphid prey detection rates between sampling dates in other beetles and spiders, this was not the case for staphylinids, T. quadristriatus and N. biguttatus (fig. 2a).

Fig. 2. (a) Aphid numbers (Sitobion avenae; 18 replicate samples per date) (◊) and aphid DNA detection rates (%) (●) and (b) aphid parasitism rates (%) (◊) and parasitoid DNA detection rates (%) (●) in arthropod predators sampled at four dates between 24 May and 3 July 2005 within a winter wheat field near Warwick, UK. Prey DNA detection rates in spiders were down-weighted to allow for better comparison with beetle predators. Different letters denote non-overlapping confidence limits in DNA detection rates.
Table 1. Percentages of arthropod predators testing positive for DNA of Sitobion avenae (aphid DNA) and parasitoid DNA. Predators were collected between 24 May and 3 July 2005 in a winter wheat field near Warwick, UK. For spiders, down-weighted detection rates are provided. Lower (lCL) and upper (uCL) 95% tilting confidence limit; different letters denote non-overlapping confidence limit between taxa listed within the same column.
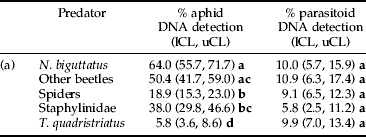
To estimate the average predation on unparasitized S. avenae over the whole season, only predators which tested positive for aphid DNA were considered, excluding those that also contained parasitoid DNA. This reduced S. avenae detection rates by approximately 30% in spiders and T. quadristriatus, as well as by approximately 10% in the other three predator groups. Of all predators where solely aphid DNA was detected, N. biguttatus, other beetles, spiders, rove beetles and T. quadristriatus accounted for 32.4%, 23.6%, 20.1%, 18.1% and 5.8%, respectively.
Predation on parasitoids
Almost all parasitoid species targeted by the molecular assay were found to be consumed by the five predator groups when the data from the four sampling dates were pooled (table 2). Exceptions were T. quadristriatus, in which no A. ervi DNA was found, staphylinids (no D. carpenteri) and other beetles (no A. picipes).
Table 2. Percentages of arthropod predators testing positive for DNA of the five primary parasitoid species: Aphidius ervi (Ae), A. picipes (Ap), A. rhopalosiphi (Ar), A. uzbekistanicus (Au) and Ephedrus plagiator (Ep), as well as the hyperparasitoid Dendrocerus carpenteri (Dc). For spiders, down-weighted detection rates are provided. Predators were collected between 24 May and 3 July 2005 in a winter wheat field near Warwick, UK. Lower (lCL) and upper (uCL) 95% tilting confidence limit; different letters denote non-overlapping confidence limit between taxa listed within the same line.

Parasitism rates by the six parasitoid species, detected using diagnostic PCR, ranged from 11.3% to 21.4% between June and July (fig. 2b). Although no parasitized aphids were found in the wheat samples taken on 24 May, 5.6%, 9.8% and 12.5% of N. biguttatus, other beetles (mostly carabids) and spiders (detection rates down-weighted), respectively, tested positive for parasitoid DNA (fig. 2b). Overall, no significant differences in parasitoid DNA detection rates between the five predator taxa were found (table 1). The same was true for parasitoid detection over time, except for T. quadristriatus, where the proportion of parasitoid positive beetles was significantly higher on 8 June compared to 3 July (fig. 2b).
From all other beetles, spiders, T. quadristriatus, Staphylinidae and N. biguttatus assayed in the present study, 8.6%, 7.9%, 5.8%, 3.5% and 2.6%, respectively, contained parasitoid DNA in the absence of aphid DNA. This indicates that, aside from N. biguttatus (26.0%) and spiders (41.8%), in all other predator taxa, more than 50% of the parasitoid DNA stems from direct predation on adult parasitoids (T. quadristriatus 86.9%, Staphylinidae 60.3%, other beetles 53.2%). The rate of predators containing solely parasitoid DNA changed between sampling dates; it was 13.8%, 7.6%, 8.7% and 2.6% for 24 May, 8 June, 21 June and 3 July, respectively.
Among the parasitoid prey, E. plagiator was detected in higher proportions than other parasitoid species in spiders, T. quadristriatus and N. biguttatus (except D. carpenteri in N. biguttatus) (table 2). In spiders, rates of detection of the hyperparasitoid D. carpenteri were lower compared to those of the primary aphid parasitoids (except for A. ervi) (table 2).
More T. quadristriatus tested positive for parasitoids than for aphids, and spiders were characterized by a comparatively high parasitoid:aphid prey detection ratio (approx. 1:2) (table 1). Within the other predator groups, significantly more predators tested positive for S. avenae than for parasitoid prey (table 1). Moreover, 29.7% of T. quadristriatus and 31.8% of spiders which contained aphid DNA also tested positive for parasitoid prey. In the other three predator taxa/groups, however, these proportions ranged only between 9.6% and 12.5%.
Discussion
The results show that generalist predators are tightly connected to both cereal aphids and their parasitoids, although the levels at which these prey were taken varied significantly among predator taxa. Aphid prey detection, in the absence of parasitoid DNA, ranged between 14% and 32% in N. biguttatus, other beetles, rove beetles and spiders. Taking these percentages as a proxy for aphid consumption, all four taxa seem to regularly feed on aphids. Trechus quadristriatus, albeit representing 31.3% of all predators caught, accounted for only 5.8% of predators in which solely aphid DNA could be detected. By contrast, this carabid species made up 49.2% of cases where only parasitoid DNA could be detected, followed by spiders, which made up for 27.1% of all parasitoid-positive predators. This conservative surrogate for predation on adult parasitoids indicates that these two predator taxa, in particular, might be capable of diminishing the adult parasitoid population. In both of these predator taxa, approximately one third of the beetles and spiders which contained aphid DNA also tested positive for parasitoid prey. In the other three predator taxa/groups, however, these proportions ranged only between 9.6% and 12.5%, indicating that T. quadristriatus and spiders were not only the main predators of adult parasitoids but also responsible for the majority of coincidental intraguild predation on parasitoids.
During the four sampling dates, on average 17.2% of the collected aphids were parasitized and one would expect a similar percentage of parasitized prey within the predators if random feeding occurs. Therefore, our data suggest that, in both T. quadristriatus and spiders, either the consumption of parasitized aphids (i.e. coincidental IGP) was disproportionately high or many aphids and parasitoids were consumed in separate predation events and the DNA from both prey detected simultaneously in the guts of these predators. The latter would mean that direct predation on adult parasitoids in these groups is underestimated in our results. In both cases, T. quadristriatus and perhaps also spiders are likely to have reduced control of aphids by parasitoids, while simultaneously assisting aphid control through predation on these pests. The potential of generalist predators to disrupt parasitoid top-down control of aphids has already been shown experimentally by the carabid Pterostichus melanarius (Illiger) preying on A. ervi (Snyder & Ives, Reference Snyder and Ives2001), although it was not known whether this was caused via feeding on mummified aphids or on other parasitoid developmental stages.
Interestingly, predators not only regularly consumed both parasitoids and their aphid hosts, but consumption of specific parasitoid species varied between predator species and over time. For example, predation rates by spiders, T. quadristriatus and N. biguttatus on the primary parasitoid E. plagiator were higher than on other primary parasitoids that showed similar or even higher rates of aphid parasitism (Traugott et al., Reference Traugott, Bell, Broad, Powell, van Veen, Vollhardt and Symondson2008). Examining functional traits of both prey and predators may help to elucidate the mechanisms which drive these specific aphid-parasitoid-predator feeding interactions. For example, E. plagiator shows long developmental times and long adult lives compared to the other primary parasitoid species (Ruggle & Holst, Reference Ruggle and Holst1994). This may have increased its exposure to IGP compared to the other parasitoid species.
IGP is common in nature and can have a large influence on the trophic structuring of communities (Vance-Chalcraft et al., Reference Vance-Chalcraft, Rosenheim, Vonesh, Osenberg and Sih2007). In the case of aphid-parasitoid communities, it has been hypothesized that IGP drives fluctuations in parasitoid populations and can substantially alter the effectiveness of parasitoids to control herbivores (Brodeur & Rosenheim, Reference Brodeur and Rosenheim2000). The current data predict that direct IGP on aphid parasitoids is a common phenomenon in generalist predators, as more than 50% of the parasitoid DNA stems from direct predation on adult parasitoids. It is possible that some predators consumed parasitized R. padi and M. dirhodum and, as the aphid primers employed in our multiplex PCR were specific to S. avenae, only the DNA of the parasitoid would be detectable in these cases. However, as the densities of these two aphid species were much smaller compared to those of S. avenae (together ∼4% of all aphids collected), coincidental IGP events involving R. padi and M. dirhodum are unlikely to have significantly affected our estimate of direct predation on adult parasitoids. Interestingly, the predation pressure on parasitoid adults seemed to be higher at the first two sampling dates, when two-thirds of the parasitoid-positive predators contained exclusively parasitoid DNA, compared to 21 June and 3 July, when this rate dropped to one-third. The rate of predators testing positive exclusively for aphid DNA, however, was largely unaffected by sampling date, ranging between 75% and 87%. These high rates of direct IGP on aphid parasitoids are surprising, contrasting with earlier studies which suggested that strong IGP on aphid parasitoids is unlikely to occur (Cardinale et al., Reference Cardinale, Harvey, Gross and Ives2003; Schmidt et al., Reference Schmidt, Lauer, Purtauf, Thies, Schaefer and Tscharntke2003). The dearth of data on this type of IGP, however, is probably a reflection of the lack of suitable techniques in the past for detecting such links (but see Nakamura & Nakamura, Reference Nakamura and Nakamura1977), which can have important implications for both host and parasitoid populations. Nevertheless, high rates of direct predation on parasitoid adults have been considered in earlier work as well; for example, Völkl & Kraus (Reference Völkl and Kraus1996) calculated that 50% of female Pauesia unilachni (Hymenoptera: Braconidae), parasitoids of the grey pine aphid Schizolachnus pineti (Homoptera: Lachnidae), were expected to be caught by spiders during their first 24-h foraging period, which significantly affected parasitoid fitness and aphid control.
Our data suggest that coincidental IGP is common too, especially by spiders where parasitized aphids might constitute a significant proportion of their aphid prey (∼30%). This highlights the need to consider coincidental IGP as a potentially significant driver, affecting both aphid and parasitoid population dynamics. As feeding experiments have shown that it is more difficult to detect the DNA of early stage parasitoids when they are consumed with their aphid hosts (Traugott & Symondson, Reference Traugott and Symondson2008), consumption of these early stage parasitoids, therefore, was probably underestimated. Moreover, behavioural aspects can increase predation rates on aphids as well. Generalist predators have been shown to aggregate at places where aphid densities are high, which leads to enhanced predation rates on parasitized aphids and in turn negatively affects parasitoid adult emergence (Chacon & Heimpel, Reference Chacon and Heimpel2010).
The trophic data presented here indicate which predators show frequent trophic linking and, hence, are likely to affect the population dynamics of aphids and parasitoids. By examining these feeding interactions at several time points during aphid population development, temporal changes in the frequency of specific trophic links could be recorded. Manipulative experiments, in field cages, could now be used to determine how these trophic links, including their temporal dynamics, directly and indirectly affect the population dynamics of aphids and parasitoids. For example, the current data suggest that T. quadristriatus and linyphiid spiders act as important intraguild predators of both adult and immature aphid parasitoids during the build up of the aphid population. This IGP should negatively affect control of aphids by parasitoids, a hypothesis which can be tested in such experiments. Combining manipulative experimentation with molecular analysis of predation may also reveal why, in diverse predator communities, IGP does not necessarily disrupt herbivore control (e.g. Snyder et al., Reference Snyder, Finke and Snyder2008).
No aphids were found during field sampling at the end of May, yet 28% of spiders and beetles collected at that first sampling date tested positive for aphid DNA. This indicates that the number of tillers inspected for aphids at this time was clearly too small to record the first aphids invading the field or that the aphids were largely being consumed by predators as quickly as they arrived. In previous studies, a similar phenomenon was found, with disproportionately high rates of aphid consumption by spiders before aphid numbers began to increase later on in the season (Harwood et al., Reference Harwood, Sunderland and Symondson2004, Reference Harwood, Desneux, Yoo, Rowley, Greenstone, Obrycki and O'Neil2007; Kuusk et al., Reference Kuusk, Cassel-Lundhagen, Kvarnheden and Ekbom2008). It has been shown that predation on aphids early in the season can significantly decrease cereal aphid densities and delay their exponential increase (Edwards et al., Reference Edwards, Sunderland and George1979; Chiverton, Reference Chiverton1987). High early-season rates of predation on aphids in the current study, therefore, may have provided a significant contribution to aphid control. However, besides feeding on aphids, 7.5% of the predators collected at the end of May had also consumed aphid parasitoids. This potentially weakens aphid control by parasitoids during their establishment phase, and more work is needed on the temporal affects of IGP on aphid control in future. Without molecular methods, these important early season interactions between generalist predators, aphids and their parasitoids would have been overlooked.
Although aphid densities increased significantly over time, the frequency of aphid DNA detection was not correlated with aphid availability in most predator groups. Similarly, detection rates of parasitoid prey were not correlated with the percentage of parasitized aphids available. Thus, factors other than the abundance of aphids might also have driven predation rates on aphids and parasitoids by generalist predators. For example, recent work has shown that the availability of alternative prey affects the level of predation on aphids by spiders and carabid beetles (Harwood et al., Reference Harwood, Sunderland and Symondson2004; von Berg et al., Reference von Berg, Thies, Tscharntke and Scheu2009; Kuusk & Ekbom, Reference Kuusk and Ekbom2010), a phenomenon which might have affected predation on parasitoids as well. The densities of the predators changed significantly between the four sampling dates (fig. 1), being 1.5 to 3 times higher in July than on all earlier dates. Taking this numerical response into account, the actual predation rate (number of prey taken per predator) for both aphid and parasitoid prey probably increased significantly towards the aphids' population maximum, although relative aphid and parasitoid DNA detection rates were largely unaffected by sampling date. Note, however, that the increase in predator densities was largely affected by T. quadristriatus, a species which was found to prey less voraciously on aphids but which showed considerable detection frequencies of parasitoid DNA. Less abundant predators, such as N. biguttatus, staphylinids and other beetles, that showed high rates of aphid consumption throughout the aphid season, are likely to deliver a similar or even higher biocontrol service than the more abundant spiders and T. quadristriatus.
Predation rates by different taxa, measured using PCR, should always be compared with caution, as prey DNA detection success can vary significantly between different species (Greenstone et al., Reference Greenstone, Rowley, Weber, Payton and Hawthorne2007; King et al., Reference King, Read, Traugott and Symondson2008). To adjust for inter-specific variation in DNA detection rates, data from feeding experiments can be used (Szendrei et al., Reference Szendrei, Greenstone, Payton and Weber2010). However, adjusting field-derived molecular gut content data by feeding experiments is only feasible when assessing limited numbers of predator-prey links (e.g. Szendrei et al., Reference Szendrei, Greenstone, Payton and Weber2010) and would require an impractically large amount of work to adjust feeding networks such as the current one. Thus, to allow for a more realistic comparison of prey found in predatory beetles and spiders in the present study, we down-weighted prey detection rates of spiders (mostly linyphiids), as previous studies have shown that post-feeding prey detection success is significantly higher in spiders than in beetles (e.g. Sheppard et al., Reference Sheppard, Bell, Sunderland, Fenlon, Skervin and Symondson2005; Traugott & Symondson, Reference Traugott and Symondson2008). This adjustment showed that the role of spiders as predators of both aphids and parasitoids is comparable to that of the mainly small- and medium-sized beetle predators. Other unmeasured or unmeasurable parameters, such as feeding frequency and prey developmental stage, can affect prey DNA detection success too (e.g. all developmental stages may be present in the field simultaneously) (Traugott & Symondson, Reference Traugott and Symondson2008). Hence, although temporal changes within a particular trophic link can be analyzed easily, it is generally more difficult to compare prey detection rates between different taxa. The current data provide, nevertheless, new insights into the occurrence and frequency of direct and ‘hidden’ (parasitized prey) feeding interactions within an aphid-parasitoid-generalist predator community under natural conditions, with major implications for future work.
Acknowledgements
This study was funded by a Marie Curie Intra European Fellowship (no. 515216 P-P INTERACTIONS) granted to MT and held at Cardiff University with WOCS and a BBSRC grant no. 72/D19632 entitled ‘Utilizing ecological profiling to evaluate the significance of predator biodiversity for sustainable pest regulation’ held by WOCS at Cardiff University jointly with David Skirvin at HRI-Wellesbourne. We thank Lidija Kravar-Garde, Kelly Reynolds, Samuel Sheppard and David Skirvin for help during field work. We are also grateful to Corinna Wallinger and two anonymous referees for their suggestions which improved this paper and to Gilg Seeber for statistical advice.
Supplementary material
The online appendix and tables can be viewed at http://journals.combridge.org/ber.






