Introduction
Codling moth Cydia pomonella (L.) (Lepidoptera: Tortricidae) is a major insect pest of deciduous fruits in temperate areas worldwide. It infests the fruits of several cultivated tree species, mainly apple Malus domestica Borkh (Rosaceae), pear Pyrus spp. (Rosaceae), quince Cydonia oblonga Mill (Rosaceae) and walnut Juglans spp. (Juglandaceae) (Shel'Deshova, Reference Shel'Deshova1967; Barnes, Reference Barnes, van der Geest and Evenhuis1991). The phenology of the species is quite variable with one to five generations per year. Temperature and photoperiod seem to be the major driving forces of this variation (Riedl & Croft, Reference Riedl and Croft1978), but pleiotropic effects of costly insecticide resistance mechanisms (Boivin et al., Reference Boivin, Bouvier, Beslay and Sauphanor2004) and adaptation to host-plants (Phillips & Barnes, Reference Phillips and Barnes1975) are also involved. Codling moth has two traits which are interesting from both a theoretical and a practical point of view. First, it has developed resistance to various classes of chemical insecticides in several parts of the world (e.g. Europe: Sauphanor et al., Reference Sauphanor, Bouvier and Brosse1998; USA: Knight et al., Reference Knight, Brunner and Alston1994, South America: Fuentes-Contreras et al., Reference Fuentes-Contreras, Reyes, Barros and Sauphanor2007). This has been attributed to the intensive chemical control against the pest. In some cases, insecticide treatments are the main force affecting the genetic structure and regulating the dynamics of codling moth populations (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007). Second, the insect has achieved an almost globally distribution, presumably due to its potential to adapt in different environments. Reasons for this success have been reported by Thaler et al. (Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008). Briefly, these are the re-emergence of two refugial haplotype clades with a long history of independent evolution leading to a diverse genetic heritage, the plasticity of the species to survive and reproduce under different climatic conditions and host-trees, the development of insecticide resistance and, lastly, the human activities which facilitate dispersal and intermixing of different genotypes and spread of resistance to insecticides.
These characteristics make codling moth a good model for studying ecological questions related to migration and gene flow among neighbouring or geographical distant populations. To shed light on dispersal process, several studies have analysed the structure of codling moth populations using either allozyme (Buès & Toubon, Reference Buès and Toubon1992; Buès et al., Reference Buès, Toubon and Poitout1995) or more recently AFLP (Timm et al., Reference Timm, Geertsema and Warnich2006; Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008) and microsatellite markers (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007; Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramírez2008; Chen & Dorn, Reference Chen and Dorn2010). Studies on temporal and spatial variation at molecular level could provide information for the pest fitness and success (Armstorng & Wratten, Reference Armstrong, Wratten, Symondson and Liddell1996), and they aid also the development of Integrated Pest Management strategies (Denholm & Rowland, Reference Denholm and Rowland1992).
Theoretical aspects of spatial and temporal genetic patterns in relation to gene flow have been reviewed by Baur & Schmid (Reference Baur, Schmid and Gaston1996) and Roderick (Reference Roderick1996). The strength of gene-flow among insect populations may be affected by the dispersal ability (distance and tendency for migration/dispersal) and limited gene flow is often observed in populations of relatively sedentary species. However, other biological traits, such as population increase and host-plant specialisation, and ecological factors related to habitat (landscape characteristics, geographical barriers, elevation, climatic constraints and host-plant isolation) can also often modulate gene-flow (Peterson & Denno, Reference Peterson and Denno1998a,Reference Peterson, Denno, Mopper and Straussb; Bohonak, Reference Bohonak1999; Loxdale & Lushai, Reference Loxdale, Lushai, Woiwod, Reynolds and Thomas2001). Knowledge about dispersal in insects and gene flow among their populations has been obtained mainly from two classes of methods. Directs methods, such as mark-release-recapture (MRR), can provide information on the contemporary pattern of migration, but they have some practical difficulties and limitations (e.g. dilution at large spatial distances). Also, they cannot estimate the contribution of emigrants to the local populations, as migration is not always translated into successful colonisation. Indirect methods use various genetic markers, and the required data (allele and genotype frequencies) for the estimation of gene-flow and migration rates are relatively easy to gather. These methods can provide long-term indirect estimates of gene flow, averaged over numerous generations and integrated geographically over many populations; but patterns of contemporary movements of individuals or propagules can be also addressed. Such information can assist in understanding the dynamics of pest expansion (Roderick, Reference Roderick1996; Peterson & Denno, Reference Peterson and Denno1998a,Reference Peterson, Denno, Mopper and Straussb; Bohonak, Reference Bohonak1999; Loxdale & Lushai, Reference Loxdale, Lushai, Woiwod, Reynolds and Thomas2001; Manel et al., Reference Manel, Gaggiotti and Waples2005).
Cydia pomonella is traditionally considered as a sedentary species, and MRR experiments with males have shown that the majority disperse within 60–80 m, although a small proportion of males is able to fly up to several kilometres (Mani & Wildbolz, Reference Mani and Wildbolz1977; Keil et al., Reference Keil, Gu and Dorn2001). Laboratory experiments confirmed the results of MRR and showed also similar flight capacity for males and females (Schumacher et al., Reference Schumacher, Weyeneth, Weber and Dorn1997). This variation in dispersal is considered adaptive, as it provides opportunities for survival in cases of habitat deterioration (Schumacher et al., Reference Schumacher, Weyeneth, Weber and Dorn1997; Keil et al., Reference Keil, Gu and Dorn2001) and, along with transportation of infested material (fruits, harvest bins) (Hibgee et al., Reference Higbee, Calkins and Temple2001), might have important implications on gene flow between distant populations. Studies with allozyme (Buès et al., Reference Buès, Toubon and Poitout1995) and DNA (Timm et al., Reference Timm, Geertsema and Warnich2006; Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007; Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramírez2008; Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008; Chen & Dorn, Reference Chen and Dorn2010) markers on populations from various regions of the world have revealed variable results. Despite some evidence of genetic differentiation among geographically close populations with different kinds of markers, the general pattern is that genetic differentiation among distant population remains globally low (reviewed by Franck & Timm, Reference Franck and Timm2010).
In contrast to codling moth from western and Central Europe, there is no study on the genetic structure of populations from Greece. Codling moth in Greece is a major pest in apple orchards, and its control is based almost exclusively on chemical insecticides. Apple cultivation is distributed throughout mainland Greece at locations with different climatic conditions and landscape characteristics; and often apple and other fruits, hosts of codling moth, are cultivated in the same areas. These diversified habitats of codling moth may provide opportunities for population subdivisions and locally adapted populations. In addition, there is no information about the genetic similarity of codling moth populations from Greece with those from other European countries, such as France, where diversified habitats also exist and similar chemical control programs are applied. Comparative studies among populations from different countries can aid in the assessment of gene-flow over large spatial scales and shed light on the possible role of human activities, such as commerce, in pest dispersal.
Therefore, we used 11 microsatellites, which are highly polymoprhic codominant markers suitable for population genetic analysis (Behura, Reference Behura2006), to assess the genetic structure of codling moth populations from different hosts and locations in Greece and southern France. An additional aim was to infer about potential gene flow over various geographical scales and adaptation to different hosts or environmental conditions. Such data might also be useful for the improvement of codling moth control programs, as they are often affected by the dispersal of the pest genotypes among orchards and regions.
Material and methods
Insect material
Fifteen samples of full grown diapausing codling moth larvae were collected from apple, walnut and pear orchards from various localities in Greece, with different altitudes (7–1122 m), covering a south-north cline. In addition, samples from the above hosts were collected in localities of southern France (altitude range 20–36 m). Larvae were collected using corrugated cardboard traps, and microsatellite genotyping analysis was performed on the emerging adults (table 1, fig. 1).

Fig. 1. Sampling sites and host-trees in Greece and France. 1, Alexandria (apple); 2, Agia (apple, walnut); 3, Zagora (apple), Lehonia (pear), Drakia (walnut); 4, Argos (pear); 5, Tripoli (apple); 6, St-Marcelin (walnut); 7, Valence (apple, pear, walnut); 8, Avignon (apple, pear).
Table 1. Insect material. Orchard location and number of codling moth larvae (N) collected.

* NGR, north Greece; CGR, central Greece; SGR, south Greece; SFR, south France.
Microsatellite genotyping analysis
Total DNA was extracted from one leg of each moth using 200 μl of 10% Chelex 100 (Biorad, San Francisco, CA, USA) resin solution (Walsh et al., Reference Walsh, Metzger and Higuchi1991). Eleven microsatellite loci (Cp3.180, Cp3.169, Cp2.129, Cp1.62, Cp5.24, Cp5.M, Cp4.S, Cp6.46, Cp2.131, Cp4.129, Cp6.32) (Franck et al., Reference Franck, Guérin, Loiseau and Sauphanor2005, Reference Franck, Ricci, Klein, Olivares, Simon, Cornuet and Lavigne2011) were examined in a total of 413 moths. Details on microsatellite loci amplification, analysis and visualization are presented in previous papers (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007, Reference Franck, Ricci, Klein, Olivares, Simon, Cornuet and Lavigne2011).
Basic statistics, Hardy-Weinberg equilibrium and linkage disequilibrium
We examined the data for presence of null alleles using Microchecker version 2.2.3 (van Oosterhout et al., Reference van Oosterhout, Hutchinson, Wills and Shipley2004). In addition, we used the method of Brookfield (Reference Brookfield1996) as implemented in GENEPOP version 4.0 (Rousset, Reference Rousset2008) to calculate the frequency of null alleles (A n). Due to the low frequencies of null alleles found (see results), data from all 11 loci were used in the subsequent analyses.
Allele frequencies, number of alleles per locus, observed (H O) and expected (H E) heterozygosity, as well as inbreeding coefficient (F IS), were calculated using GENEPOP version 4.0. Allelic richness (Rs, number of alleles independent of sample size) was calculated using FSTAT version 2.9.3.2 (Goudet, Reference Goudet1995). Deviation from Hardy-Weinberg equilibrium (HWE) at each locus and across all loci (U test and multisample score test, respectively; Raymond & Rousset, Reference Raymond and Rousset1995) and linkage disequilibrium between pairs of microsatellite loci (G log-likelihood based exact test; Goudet et al., Reference Goudet, Raymond, de Meeüs and Rousset1996) were tested using GENEPOP version 4.0.
Changes in the population effective sizes were examined using the software BOTTLENECK version 1.2.02 (Cornuet & Luikart, Reference Cornuet and Luikart1996). The observed gene diversities were compared to their expected values from the number of alleles in the samples under the assumption of mutation-migration-drift equilibrium. Gene diversities at the mutation-migration-drift equilibrium were computed for the two-phase mutation model (TPM; with 95% single-step mutations and 5% multiple-step mutations), which is considered appropriate for microsatellites (Piry et al., Reference Piry, Luikart and Cornuet1999). A significant excess of observed gene diversity relative to the expected gene diversity at the equilibrium may indicate a population size reduction, while a deficit may indicate that the population is growing. Significances were tested with two-tailed Wilcoxon's signed-rank test (Piry et al., Reference Piry, Luikart and Cornuet1999).
AMOVA and F ST analysis
Population structure was assessed by calculating multilocus F ST values (Weir & Cockerham, Reference Weir and Cockerham1984) for pairwise comparisons of samples using ARLEQUIN version 3.11 (Schneider et al., Reference Schneider, Roessli and Excoffier2000). Significance (P-value) was assessed with 10,000 permutations of diploid multilocus genotypes between samples. The structure of the data was also investigated by analysis of molecular variance (AMOVA) with ARLEQUIN version 3.11 using a permutation nonparametric approach (1000 permutations) to test for the significance of fixation indices (Excoffier et al., Reference Excoffier, Smouse and Quattro1992). The partitioning of variance between groups, i.e. Greek and French samples, or three groups according to host-trees (apple, pear, walnut), among samples within groups, and within samples was examined.
To test for isolation by distance (IBD) among codling moth samples a regression analysis of the linearised F ST transformation (F ST/(1–F ST)) onto the logarithm transformation of the geographical distance was performed (Rousset, Reference Rousset1997) using GENEPOP version 4.0. The significance of this regression was based on Mantel's test (Mantel, Reference Mantel1967) using 1000 permutations of pair of samples.
Bayesian clustering and genetic distance analyses
Bayesian analysis implemented in STRUCTURE version 2.2 (Pritchard et al., Reference Pritchard, Stephens and Donnelly2000) was used to infer the number of K unknown genetic clusters in which the sampled multilocus genotypes can be split. The method also assigns a probability that the individuals belong to a certain cluster or to more than one cluster if they are admixed. We used both of the two ancestry models that STRUCTURE provides (admixture, each individual draws some fraction of its genome from each of the K populations; and no admixture, individuals are discretely from one population or another) without any prior information about the samples (i.e. sampling region, host-plant). In addition, two models for the allele frequencies (independent allele frequency, IAF; correlated allele frequency, CAF) were used for each of the two ancestry models. The IAF model assumes that the allele frequencies in each population are independent draws from a distribution, while the CAF model assumes that allele frequencies in the different populations are likely to be similar (probably due to migration or shared ancestry). In each case, we ran ten simulations (to check the consistency of our data) of 100,000 iterations following a burn-in period of 50,000 iterations for each K-value, and with values of K from 1 to 10. The pointers provided by Pritchard et al. (2000), Garnier et al. (Reference Garnier, Alibert, Audiot, Prieur and Rasplus2004) and Evanno et al. (Reference Evanno, Regnaut and Goudet2005) was used to select the K-value showing the best subdivision of our data. We examined the increase pattern of the estimated posterior probability of the data (mean LnPPD over ten runs divided by standard deviation) with K and the distribution of ΔLnPPD values (mean of the absolute values of the rate of change of LnPPD averaged over ten runs divided by standard deviation). The modal value of this distribution is located at the real K and its height is used as an indicator of the strength of the signal detected by STRUCTURE (Evanno et al., Reference Evanno, Regnaut and Goudet2005).
To verify the K-value found using STRUCTURE, we also analyse the multilocus genotypes with BAPS version 5.3 (Corander et al., Reference Corander, Marttinen, Sirén and Tang2008). In particular, we run a mixture analysis with the multilocus genotypes pre-assigned to the 15 samples of codling moth examined. Ten runs for each K=2–10 were performed.
In addition, the GeneClass2 software (Piry et al., Reference Piry, Alapetite, Cornuet, Paetkau, Baudouin and Estoup2004) was used for population assignment and exclusion tests, including the calculation of probability of origin for each individual moth. We used the Bayesian method of Rannala & Mountain (Reference Rannala and Mountain1997) as computation criterion. The probability for the assignment of the individuals to a given population was computed using the simulation algorithm of Paetkau et al. (Reference Paetkau, Slade, Burden and Estoup2004) for Monte-Carlo re-sampling, and 1000 simulations were applied with an error rate of 0.01 for type I errors (assignment threshold probability). Assignment tests were also run without probability computation (without Monte-Carlo re-sampling). The score for each individual to the relative populations was calculated and the assignment threshold of scores was 0.05.
To further investigate the genetic relationship between populations, a neighbour joining (NJ) tree based on the allele shared distance (DAS) (Chakraborty & Jin, Reference Chakraborty, Jin, Pena, Chakraborty, Epplen and Jeffreys1993) was constructed using the software POPULATIONS version 1.1.28 (http://bioinformatics.org/~tryphon/populations/). DAS distance counts the number of different alleles between multilocus genotypes. Bootstrap values were calculated by resampling loci, and are presented as percentages over 1000 replications. The tree was constructed using the software MEGA version 4.01 (Tamura et al., Reference Tamura, Dudley, Nei and Kumar2007). In addition, we used GENALEX 6.1 software (Peakall & Smouse, Reference Peakall and Smouse2006) to calculated pairwise PhiPT distance values (an analogue of F ST) in the examined populations. A principal components analysis (PCA) was performed using the matrix of the PhiPT values.
Results
Basic statistics, Hardy-Weinberg equilibrium and linkage disequilibrium
In total, 258 individuals from nine Greek localities and 155 individuals from six French localities were analysed. No evidence for null allele was found in the 11 loci using MICROCHECKER software. In addition, low frequencies of null alleles were calculated using GENEPOP, which ranged from 0.000 to 0.179 for each locus with mean values over all loci for the 15 populations 0.003–0.036. The number of alleles ranged from 3 to 29 and allelic richness from 2.2 to 15.5 (table 2). The mean (over all loci) number of alleles and allelic richness ranged among the Greek samples from 7.3 to 10.4 and from 6.8 to 7.8, respectively. The corresponding values for the samples from France ranged from 6.7 to 7.8 and from 6.1 to 6.6. Significant differences were found between Greek and French samples in the mean number of alleles (over all loci) (8.3 vs 7.1, U=49.5, P=0.009) and mean allelic richness (7.2 vs 6.3, U=54.0, P=0.002). By contrast, no differences were found between these two groups of samples in mean frequency of null alleles (0.024 vs 0.018, U=33.5, P=0.477), mean H EXP (0.653 vs 0.638, U=39.0, P=0.175), mean H O (0.626 vs 0.641, U=21.0, P=0.517) and mean F IS (0.04 vs –0.01, U=40.0, P=0.139) (table 3, table S1 in the supplementary material).
Table 2. Number of alleles, allelic richness and frquency of null alleles per loci over all samples from Greece (n=258) and France (n=155).
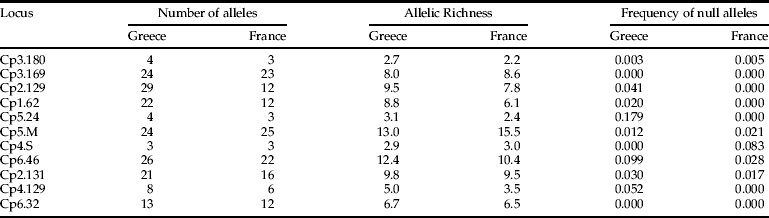
Table 3. Genetic diversity indices. Mean number of alleles (n), proportion of null alleles (A n), allelic richness (R S, number of alleles independent of sample size), heterozygosity expected (H EXP) and heterozygosity observed (H O) over all loci.

Significant deviation from HWE was found in 26 (17 in Greek and nine in French samples) out of 165 locus/population combinations. Most of these deviations were associated with heterozygote deficiency. Significant multi-locus deviations associated with heterozygote deficiency was observed in five populations, four from Greece (AgAb, AlexA, PiP, PiW) and one from France (AviP), and with heterozygote excess was indicated in one population from France (ValW) (table S1). BOTTLENECK software did not detect recent bottleneck effect in any of the populations examined. However, significant heterozygosity deficiency in the average observed gene diversity compared to the expected gene diversity under the TPM model was observed in two populations from Greece (PiP, P=0.027; TrA, P=0.001), which may suggest population expansion. Linkage disequilibrium analysis revealed 63 cases of significant associations between pairs of loci out of 825 pairwise comparisons. This value is above the expected number by chance in case of equilibrium (0.05×825=41.25). Most pairs of loci with linkage disequilibrium were observed in one population from Greece (PiW, 15 pairs) and one from France (ValA, 16 pairs).
AMOVA and F ST analysis
The global estimate of F ST over all 15 samples and the 11 microsatellite loci was 0.050 and significant different from zero (P<0.001), suggesting some genetic differentiation among samples. The hierarchical AMOVA over all loci and samples revealed small but significant variance (3.9% of the total variance, P=0.006) between groups (France vs Greece) with a global F CT=0.039 (P<0.001), although most of the variance (95.0%) was within samples. The within group variance was low (1.1%) and non-significant (P=0.074). Global F ST estimate among the Greek samples was 0.009 (P<0.001) and 0.015 (P<0.001) for the French samples. The mean value for the pair-wise comparisons between the samples from Greece and France was 0.032. The loci Cp5.24 and Cp6.32 may contribute more to the differentiation of the two groups, as they explain a high percentage of the between-group variance (13.7 and 13.3%, respectively) with F ST values of 0.151 and 0.136, respectively (table 4). The pair-wise comparisons among samples resulted in 37 significant cases out of 51 with the F ST values ranged in Greek and French samples 0.002–0.081 and 0.002–0.068, respectively (table 5). AMOVA on samples grouped by their host of origin (three groups) did not reveal significant variance in the genetic structure between the three hosts (–0.47% of the total variance, P=0.569; F CT=0.039, P=0.772).
Table 4. AMOVA results for 11 microsatellite loci. The genetic variance was partitioned between two groups (Greece and France) and among samples within these two groups.

Table 5. Bellow diagonal pairwise F ST values among populations of Cydia pomonella.

1 Abbreviations are defined in table 1. 2 NS, non significant, *P<0.05, **P<0.01 and ***P<0.001.
Mantel's test did not show significant correlation between the linearised F ST values and the geographical distance among all samples (P=0.562, df=105, r=–0.023), the Greek samples (P=0.709, df=36, r=0.105) or the French samples (P=0.699, df=15, r=–0.046).
Bayesian clustering and genetic distance analyses
Both admixture and non-admixture models showed the best solution of K to be 2 (results from the non-admixture model are presented; see fig. S1 in the supplementary material). There was a sharp increase of the LnPPD values, with K moving from 1 to 2, and then a plateau appeared to be reached. In addition, the highest (modal) value of the distribution of ΔLnPPD was observed at K=2 for both admixture and non-admixture models. It seems that splitting samples in two genetic clusters represents the optimal subdivision of the data and avoids unjustified and less informative over-splitting.
The first genetic cluster is mainly characterised by the individuals from Greece (Greek cluster) and the second by the individuals from France (French cluster) (fig. 2; the plot derived from no-admixture ancestry model and IAF model is presented). However, both of them are not pure, as there are individuals from Greece with high membership coefficient to the French cluster and vice versa. This is demonstrated also by the mean membership coefficients of the samples from Greece and France to the two clusters (table 6). The assignment of the individuals to their country of origin were further examined, and they were assigned to a single cluster (Greek or French) when their proportion of ancestry in that cluster was greater than 80%, otherwise they were considered as admixed. This empirical threshold was determined after analysing the distribution of mean ancestry coefficients of the individuals. The no-admixture ancestry model provided much better assignment of the individuals to their country of origin (Greek individuals: 77.9 and 81.0% with CAF and IAF allele frequency models, respectively; French individuals: 89.0 and 87.7% with the respective allele models) compared to the admixture model (Greek individuals: 58.5 and 21.3% with CAF and IAF allele frequency models; French individuals: 59.4 and 42.6% with the respective allele models), and this is illustrated also in the mean membership coefficient of the samples examined (table 6).

Fig. 2. Partition of genetic variation. Clustering plot of the 15 Cydia pomonella samples examined using no-admixture ancestry model and independent frequency allele model. Number of genetic clusters, K=2; Cluster 1 (Greek cluster), dark grey colour; Cluster 2 (French cluster), light grey colour. Each individual moth is represented as a vertical bar partitioned into two segments. The lengths of each segment are proportional to the estimated membership coefficients of the individual in each of the two clusters. Individuals of different samples are separated by black vertical lines. AgAa , AgAb, AlexA, AgW, ArgP, PiA, PiP, PiW and TrA are samples from Greece, AviP, AviA, MarW, ValP, ValW and ValA are samples from France; for details see table 1.
Table 6. Average membership coefficients for the two genetic clusters obtained from STRUCTURE for the 15 samples of Cydia pomonella examined.

CAF, correlated frequency allele model; IAF, independent frequency allele model.
F, French cluster; G, Greek cluster.
In agreement to the results from STRUCTURE, BAPS revealed the highest posterior probability (P=1.000) for a structure with two genetic clusters (K=2). The first cluster contained the Greek samples, and the second those from France. We also examined the assignment of the individuals in these two genetic clusters (Greek and French) using GeneClass2. The percentage of corrected classified individuals was 85.5% (98.4 and 63.9% for Greek and French individuals, respectively) using the simulation algorithm of Paetkau et al. (Reference Paetkau, Slade, Burden and Estoup2004) to compute the probability that an individual belongs to a reference population. The percentage of corrected classified individuals was increased when the assignment was based on a score test without probability computation, reaching 91.8% (92.2 and 91.0% for Greek and French individuals, respectively). In general, the assignment rates obtained by GeneClass2 were close to those obtained using STRUCTURE with the no-admixture ancestry model.
The NJ tree based on DAS (fig. 3) provided similar results with the Bayesian clustering analysis. The tree resulted in two major clusters, the first consists of the samples from France and the second those from Greece. Two major clusters were also revealed by PCA on PhiPT distance values. The separation was due to the scores of the first PCA axis, which explained 66.0% of the total variance. Variation within the two clusters was observed due to the scores of the second PCA axis, which explained a lower percentage (13.7%) of the total variance (fig. 4).

Fig. 3. Neighbour joining tree based on shared allele distance (DAS) among 15 samples of Cydia pomonella. Numbers denotes bootstrap percentages (1000 resamplings). AgAa, AgAb, AlexA, AgW, ArgP, PiA, PiP, PiW and TrA are samples from Greece; AviP, AviA, MarW, ValP, ValW and ValA are samples from France; for details see table 1.

Fig. 4. Plot of the scores of the two Axes of principal component analysis of molecular variance using the PhiPT pairwise distance matrix. The two axes explain 79.7% of the total variance. AgAa, AgAb, AlexA, AgW, ArgP, PiA, PiP, PiW and TrA are samples from Greece; AviP, AviA, MarW, ValP, ValW and ValA are samples from France; for details see table 1.
Discussion
In the present study, we investigated the genetic relationships among codling moth samples from three hosts from Greece and France, analysing 11 microsatellite loci, which is the highest number of loci compared to previous microsatellite studies on this insect species. Population structuring was found, with the most obvious case the separation of the Greek samples from those from France. However, important gene-flow was recorded among populations within countries and at a lower extent between countries.
Genetic diversity, Hardy-Weinberg equilibrium
The samples from Greece showed adequate genetic diversity, as indicated by the mean allelic richness and mean number of alleles over all loci which ranged from 6.8 to 7.8 and from 7.3 to 10.4 and the high number of alleles observed especially in seven (13–29 alleles) out of the 11 loci examined. The values are in the same order or even higher than those recorded in previous studies for codling moth populations from Europe (France, Armenia, Italy: Franck et al. (Reference Franck, Reyes, Olivares and Sauphanor2007); Switzerland: Chen & Dorn (Reference Chen and Dorn2010)) and Chile (Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramírez2008). In addition, the Greek samples showed significantly higher diversity, in terms of mean number of alleles and allelic richness, than those from France examined here. Nevertheless, the samples from both countries showed adequate molecular variance, and no recent bottlenecks were detected, suggesting a lively demographic history which is characterised by recent demographic expansions in some Greek populations.
We also observed cases of both single and multi locus significant deviations from HWE, mostly associated with heterozygote deficiency. HWE deviations have been recorded in other microsatellite studies on codling moth populations from Switzerland (Chen & Dorn, Reference Chen and Dorn2010) and Armenia (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007) but not in populations from France, Italy (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007) and Chile (Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramírez2008). In various microsatellite studies on lepidopteran species, including codling moth (Chen & Dorn, Reference Chen and Dorn2010), the significant departures from HWE (heterozygote deficiency) have been attributed to the presence of null alleles, whose frequency seems to be greater in Lepidoptera than in other insect orders (Meglécz & Solignac, Reference Meglécz and Solignac1998; Keyghobadi et al., Reference Keyghobadi, Roland and Strobeck1999; Meglécz et al., Reference Meglécz, Petenian, Danchin, Coeur, D'Acier, Rasplus and Faure2004; Endersby, et al., Reference Endersby, McKechnie, Ridland and Weeks2006; Orsini et al., Reference Orsini, Wheat, Haag, Kvist, Frilander and Hanski2008). In our study, MICRO-CHECKER did not find evidence for null allele at the 11 loci examined, and frequencies of null allele according to the method of Brookfield (Reference Brookfield1996) were generally lower than that reported in previous microsatellite studies on codling moth (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007; Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramírez2008; Chen & Dorn, Reference Chen and Dorn2010). Therefore, other factors should be responsible for the deviations from HWE observed here. In other insect orders, such as aphids, significant heterozygote deficiency in sexual populations have been attributed to selection, Wahlund effect, inbreeding and other population effects (e.g. Delmotte et al., Reference Delmotte, Leterme, Gauthier, Rispe and Simon2002; Fenton et al., Reference Fenton, Malloch, Navajas, Hillier and Birch2003). Selection of insecticide resistant genotypes due to intensive chemical control and their inbreeding could account for the HWE deviations (heterozygote deficiency) observed in the present study. The cases of linkage disequilibrium found support the possibility of inbreeding and non-random mating. Codling moth is a sedentary species, and this behaviour might enhance the inbred of selected resistant genotypes. However, Wahlund effect of sampling from distinct gene pools in the same population cannot be excluded. In support of this, Bayesian clustering analysis showed that some populations contain members of both genetic clusters into which the multilocus genotypes were split (see fig. 2 and below).
Genetic structure, gene-flow
Both Bayesian clustering and genetic distance analyses revealed a genetic differentiation between the samples from Greece and those from southern France. This was also supported by AMOVA and F ST analysis and somehow by the significant differences in some of the genetic diversity indices examined. Part of the genetic differentiation between the French and the Greek samples could be attributed to historical events. An example might be a different level of admixture in these countries of the two geographically isolated mitochondrial genotype clades which were split during the Pleistocene but come into contact after disappearance of the geographic barriers due to climatic changes (Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008). Geographic distance might then have enhanced genetic differentiation through genetic drift or local adaptation phenomena. Selection forces related to environment and to pest management practices can shape the genetic structure of local populations (Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008). Recently, Franck et al. (Reference Franck, Reyes, Olivares and Sauphanor2007) have shown that codling moth populations from France and other countries were mainly structured according to the history of insecticide applications. In support of this, the kdr allele, which confers resistance to pyrethroids, has been recorded often in France, but not yet in numerous samples from Greece, including those examined in the present study (Voudouris et al., Reference Voudouris, Sauphanor, Franck, Reyes, Mamuris, Tsitsipis, Vontas and Margaritopoulos2011).
Although we sampled codling moth from diverse habitats in Greece, in terms of geography, environmental conditions and host-trees, the majority of the individuals were grouped in one genetic cluster. The same was observed in the samples from southern France examined here. Supporting evidence for the lack of substantial population structuring within the two countries compared to the genetic variation between countries were provided also by the AMOVA results and the F ST values (see below). These findings are in accordance with previous studies on French populations with allozyme (Bués & Toubon, Reference Buès and Toubon1992; Buès et al., Reference Buès, Toubon and Poitout1995) or microsatellite markers (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007) and on Chilean populations with microsatellite markers (Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramírez2008). However, recent studies on populations from Central Europe (Germany, Austria, northern Italy) (Thaler et al., Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008) and South Africa (Timm et al., Reference Timm, Geertsema and Warnich2006) with AFLP markers showed high degree of genetic differentiation among populations, even at local geographical scales (e.g. <1 km in South Africa). Apart from the limited moth dispersal, variation in microclimate, habitat-specific conditions, geographic isolation, host-adaptation and pest control practices in Central Europe and relative isolation of pome fruit production areas along with absence of wild hosts in South Africa are possible reasons for these structuring patterns. Franck & Timm (Reference Franck and Timm2010) have suggested that these discrepancies among studies could also be attributed to the different affinities of the primers used. However, a recent microsatellite study on populations from Switzerland reported also important genetic differentiation at local geographic scale (even less than 10 km), which was mostly attributed to the sedentary behaviour of codling moth (Chen & Dorn, Reference Chen and Dorn2010).
The global F ST values (0.050 among all 15 samples; 0.009 among Greek samples; 0.015 among French samples; 0.032 mean value for the pair-wise comparisons between samples from Greece and France; F CT=0.039 between the groups of French and Greek samples), as well as the pair-wise values found here, are in the range of those reported in previous microsattelite studies on codling moth populations from other countries [F ST=0.066 among samples from France, Italy, Armenia and Chile, and F ST=0.006 among samples from France (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007); F ST=0.026 among samples from central Chile (Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramírez2008); F ST=0.016–0.128, with a mean 0.064 in samples from Switzerland (Chen & Dorn, Reference Chen and Dorn2010)]. In addition, early studies using allozyme loci found similar F ST values with the aforementioned ones [F ST=0.03 for samples from France (Buès et al., Reference Buès, Toubon and Poitout1995); F ST=0.066 at an intercontinental level (Pashley & Bush, Reference Pashley, Bush, Rabb and Kennedy1979)]. The F ST values found here contribute to the view of Franck & Timm (Reference Franck and Timm2010) that, globally, the codling moth populations appear to be only slightly structured among regions or countries.
It is worth also noticing that the F ST values found here for a sedentary insect are quite lower than those reported in microsatellite studies on other sedentary lepidopteran species, such as Melitaea cinxia L. (Nymphalidae) (F ST=0.200; Palo et al., Reference Palo, Varvio, Hanski and Vainola1995) and Polyommatus bellargus Rottemburg (Lycaeninae) (F ST=0.127; Harper et al., Reference Harper, Maclean and Goulson2003). By contrast, our values are closer to those reported for highly dispersive lepidopteran species in studies using microsatelitte [Plutella xylostella (L.) (Yponomeutidae), F ST=0.005 (Endersby et al., Reference Endersby, McKechnie, Ridland and Weeks2006); Helicoverpa armigera (Hübner) (Noctuidae), F ST=0.002 (Endersby et al., Reference Endersby, Hoffmann, McKechnie and Weeks2007)] or allozyme [Heliothis virescens (Fabricius) (Noctuidae), F ST=0.002 (Schneider, Reference Schneider1999); Ostrinia nubilalis (Hubner) (Crambidae) F ST=0.024 (Coates et al., Reference Coates, Sumerford and Hellmich2004)] markers. In general, dispersive species show reduced F ST values compared to those of reduced dispersal ability (reviewed by Bohonak, Reference Bohonak1999). Nevertheless, the different analyses performed in the present study (including the assignment tests; see below for discussion) come to the same conclusion, i.e. there is gene-flow at various geographic scales (mostly within the two countries) in a sedentary species, even between samples which are hundreds or thousands kilometres apart, separated by many physical barriers. It should be mentioned that sedentary behaviour has been proved also for Greek strains and populations with mark-release-recapture experiments and kinship analysis based on microsatellite markers (maximum dispersal distance ∼200 m; Voudouris et al., unpublished data). This gene-flow could not be attributed to active dispersion of codling moth individuals. The most possible explanation is passive dispersion due to human activities related to transportation over long distances of infested fruits and harvest bins, which are known to frequently carry diapausing larvae (Higbee et al., Reference Higbee, Calkins and Temple2001; Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramírez2008). However, some individuals of codling moth are able to undertake flights of several kilometres (Mani & Wildbolz, Reference Mani and Wildbolz1977; Schumacher et al., Reference Schumacher, Weyeneth, Weber and Dorn1997) and, given that commercial pome and walnut production and domestic culture of host-trees of codling moth are widespread in Greece and southern France, a ‘stepping-stone’ model of expansion could not be totally excluded. The gene-flow described by the ‘stepping-stone’ hypothesis in other sedentary species had a significant influence on the genetic homogenisation over large geographical distances (Peterson, Reference Peterson1996).
Another point of discussion is that assignment tests revealed individuals that are not assigned to their country of origin (Greece or France) and might represent hybrids between populations from these two or other countries. We cannot, however, discriminate between these two alternatives or clarify the patterns of codling moth dispersal across southern Europe, as it was not possible to analyse samples from localities spanning the geographic distance between Greece and southern France. Nevertheless, this finding could be attributed to recent immigration and supports also the view that gene-flow occur also over large geographical distances (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007; Franck & Timm, Reference Franck and Timm2010). It highlights also the impact of globalisation of commerce on the rather recent range expansion of this insect pest. Franck et al. (Reference Franck, Reyes, Olivares and Sauphanor2007) found also individuals from France and Italy that were hybrids between populations from different countries as a result of recent migration. In addition, Pashley & Bush (Reference Pashley, Bush, Rabb and Kennedy1979) suggested a European origin of Canadian and southern African populations, with New Zealand being an additional source for the latter populations. Recently, Timm et al. (Reference Timm, Geertsema and Warnich2006) come to similar conclusions after the analysis of samples from Canada, England and South Africa.
Isolation by distance
The theory of an isolation-by-distance (IBD) effect from a source area (Wright, Reference Wright1943; Slatkin, Reference Slatkin1993) predict that gene flow is most common over short geographic distances and decreases as geographic distance increases. IBD effect has been detected in several insect species (reviewed by Peterson & Denno, Reference Peterson and Denno1998a). However, we observed no significant correlation between genetic and geographic distance either among all 15 samples or among the nine Greek and among the six French samples. Other studies using microsatellite (Chen & Dorn, Reference Chen and Dorn2010) or AFLP (Timm et al., Reference Timm, Geertsema and Warnich2006; Thaler et al. Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008) markers failed also to find a significant IBD effect at various geographic scales in populations from Central Europe or from South Africa. On the other hand, two studies using microsatellites reported significant IBD effect in populations from Chile (Fuentes-Contreras et al., Reference Fuentes-Contreras, Espinoza, Lavandero and Ramírez2008) or from a larger spatial scale (populations from France, Italy Armenia and Chile) (Franck et al., Reference Franck, Reyes, Olivares and Sauphanor2007). Chen & Dorn (Reference Chen and Dorn2010) suggested that the discrepancy among studies about IBD effect could be related to the innate dispersal capacity of codling moth individuals and to anthropogenic influence on the orchard agro ecosystem. Human-mediated long distance dispersal of codling moth through transportation of infested material (reviewed by Franck & Timm, Reference Franck and Timm2010) could be another factor for the lack of IBD observed here or in other studies. Similarly, Peterson & Denno (Reference Peterson and Denno1998a) suggested that IBD was not pronounced in highly mobile insects as a result of low genetic differentiation (due to gene-flow) even between distant populations. In our case, however, the lack of samples covering the distance between France and Greece might have also contributed to the non-significant IBD effect.
Host-related genetic differences
Both Bayesian clustering and genetic distance analyses, including AMOVA and F ST statistics, failed to reveal host-associated differences among the populations examined here, and thus we have not evidence of host-race formation in codling moth from Greece or southern France. Similarly, allozyme polymorphism analysis on French populations from apple, pear, quince, walnut and apricot Prunus armeniaca L. did not reveal genetic difference related to host-tree (Buès & Toubon, Reference Buès and Toubon1992; Buès et al., Reference Buès, Toubon and Poitout1995). In addition, Timm et al. (Reference Timm, Geertsema and Warnich2006) did not find host-specific differences in AFLP patterns among samples from apple, pear and stone fruit from South Africa. However, early behavioural and physiological studies on codling moth from California reported differences between populations from apple, walnut and a stone fruit species (plum), and it was suggested that there are three well-defined host-races (Phillips & Barnes, Reference Phillips and Barnes1975). Toward this direction were the results of two recent studies using DNA markers. Thaler et al. (Reference Thaler, Brandstätter, Meraner, Chabicovski, Parson, Zelger, Dalla Via and Dallinger2008) found that individuals of a codling moth population from walnut trees in northern Italy had a different AFLP pattern than a neighbouring population from apple trees. Chen & Dorn (Reference Chen and Dorn2010) reported also genetic differentiation among populations from three host-trees (apple, apricot and walnut) in Switzerland. It seems that the genetic studies, including ours, provide contradictory results about the existence of host-adapted population or races in codling moth, and only one-third of them report host-associated differences. Chen & Dorn (Reference Chen and Dorn2010) suggested that the lack of host-associated difference in the aforementioned studies maybe be due to the use of allozyme loci (Buès & Toubon, Reference Buès and Toubon1992; Buès et al., Reference Buès, Toubon and Poitout1995), which are less polymorphic than microsatellite markers, or to the small sample sizes examined per tree species and location (Timm et al., Reference Timm, Geertsema and Warnich2006). In our study, regardless the use of more microsatellite loci than Chen & Dorn (Reference Chen and Dorn2010) (11 vs 9) and similar or higher sample sizes per host-tree, host-related genetic differences were not found. It seems, therefore, that host-adapted populations of codling moth are not quite spread; and, until now, they have been only recorded in Central Europe and California. The reason for this remains to be found, although factors related to climate and to agricultural landscape (e.g. distribution and size of the orchards of the various host-trees) might be involved. According to Barnes (Reference Barnes, van der Geest and Evenhuis1991), a host-associated race or population of codling moth could only evolve in an orchard of sufficient area of dominance.
Conclusions
The results of the present study suggested important gene-flow among samples from various regions of Greece from three hosts. The same pattern was observed in samples from southern France which were examined for comparison purposes. On the other hand, population structuring between the two countries was found. Historical events along with the relative geographic isolation, genetic drift and local adaptation phenomena might have contributed to this genetic differentiation. However, cases of gene-flow between samples from Greece and France were also detected. These results add to the growing body of evidence that codling moth populations are slightly structured at either country or global level (reviewed by Franck & Timm, Reference Franck and Timm2010), which reflects the globalisation of this insect pest. Given the sedentary nature of the species, which has been proved also for Greek strains and populations (Voudouris et al., unpublished data), the patterns of gene-flow observed here highlight the importance of agricultural practices and commerce in the passive dispersal of individuals over large distances. Our findings may have important implications for the chemical control of the pest and insecticide resistance management, as they are often affected by the dispersal of codling moth. A recent study showed that insecticide resistance is widespread in codling moth populations in the main apple-growing regions of Greece (Voudouris et al., Reference Voudouris, Sauphanor, Franck, Reyes, Mamuris, Tsitsipis, Vontas and Margaritopoulos2011). Apart from local factors related to insecticide selection pressure, this might have resulted from the gene-flow among populations recorded in the present study. Lastly, we didn't find evidence for host-adapted populations either in Greece or in France, suggesting regular gene-flow among populations from the three host-trees examined, although future research shall include samples from additional cultivated, domesticated and wild tree species.
Acknowledgements
The present study was funded by the PENED 2003 project. PENED 2003 was co-funded by the European Union (European Social Fund) by 80%, from the Hellenic State (Ministry of Development, General Secretariat for Research and Technology) by 20% (public grants) and from Hellenic Private Sector, in the framework of Measure 8.3 of Operational Programme ‘Competitiveness’ (the 3rd Community Support Programme). Partial funding was obtained by a bilateral Joint Research and Technology project between Greece and France (2006–2008) (funded by the Greek General Secretariat for Research and Technology). The study was also supported by the Postgraduate Courses ‘Biotechnology-Quality Assessment in Nutrition and the Environment’ and ‘Applications of Molecular Biology-Genetics: Diagnostic Biomarkers’ of the Department of Biochemistry and Biotechnology, University of Thessaly, Greece.
Supplementary material
The online table and figure can be viewed at http://journals.combridge.org/ber.












