Introduction
Wolbachia are gram-negative alphaproteobacteria of the order Rickettsiales and family Anaplasmataceae exhibiting symbiotic relationships with their hosts (O'Neill et al., Reference O'Neill, Giordano, Colbert, Karr and Robertson1992; Dumler et al., Reference Dumler, Barbet, Bekker, Dasch, Palmer, Ray, Rikihisa and Rurangirwa2001; Werren et al., Reference Werren, Baldo and Clark2008). They were first reported in the reproductive tissues of the mosquito Culex pipiens (Hertig & Wolbach, Reference Hertig and Wolbach1924) and, therefore, the species was named Wolbachia pipientis (Hertig, Reference Hertig1936). However, due to uncertainty about the actual taxonomic status of W. pipientis, researchers commonly refer to it simply as Wolbachia (Lo et al., Reference Lo, Paraskevopoulos, Bourtzis, O'Neill, Werren, Bordenstein and Bandi2007). Currently, based on gene sequence information, at least 13 major clades of Wolbachia known as ‘supergroups’ (A–F and H–N) have been reported (reviewed in Augustinos et al., Reference Augustinos, Santos-Garcia, Dionyssopoulou, Moreira, Papapanagiotou, Scarvelakis, Doudoumis, Ramos, Aguiar, Borges, Khadem, Latorre, Tsiamis and Bourtzis2011). All but three of these supergroups are found in arthropods, while the remaining three have so far only been found in nematodes (Casiraghi et al., Reference Casiraghi, Bordenstein, Baldo, Lo, Beninati, Wernegreen, Werren and Bandi2005; Lo et al., Reference Lo, Paraskevopoulos, Bourtzis, O'Neill, Werren, Bordenstein and Bandi2007; Haegeman et al., Reference Haegeman, Vanholme, Jacob, Vandekerckhove, Claeys, Borgonie and Gheysen2009; Augustinos et al., Reference Augustinos, Santos-Garcia, Dionyssopoulou, Moreira, Papapanagiotou, Scarvelakis, Doudoumis, Ramos, Aguiar, Borges, Khadem, Latorre, Tsiamis and Bourtzis2011). However, the great majority of arthropod Wolbachia so far described come from only two supergroups (A and B).
Wolbachia strains are globally distributed (Werren & Windsor, Reference Werren and Windsor2000) and currently these bacteria are considered the most abundant endosymbionts found in invertebrates. Wolbachia are referred to as reproductive parasites, because they induce diverse reproductive phenotypes, mainly in arthropods (Werren, Reference Werren1997; Werren et al., Reference Werren, Baldo and Clark2008). Commonly, they are associated with parthenogenesis (Weeks & Breeuwer, Reference Weeks and Breeuwer2001), phenotypic feminization of genetic males (Rousset et al., Reference Rousset, Bouchon, Pintureau, Juchault and Solignac1992), cytoplasmic incompatibility (O'Neill et al., Reference O'Neill, Giordano, Colbert, Karr and Robertson1992) and male killing (Hurst & Jiggins, Reference Hurst and Jiggins2000). Wolbachia are also thought to play important roles in speciation and local adaptation (Brucker & Bordenstein, Reference Brucker and Bordenstein2012). The importance of Wolbachia in reproductive processes depends ultimately on its prevalence, and how it is transmitted between species (Stouthamer et al., Reference Stouthamer, Breeuwer and Hurst1999). In Arthropoda, Wolbachia are believed to be primarily maternally transmitted within species (Skinner, Reference Skinner1982), but horizontal transmission also frequently occurs between species over longer evolutionary time-scales (Werren et al., Reference Werren, Zhang and Guo1995a ; Schilthuizen & Stouthamer, Reference Schilthuizen and Stouthamer1997).
About 40% of arthropod species are estimated to be infected with Wolbachia (Zug & Hammerstein, Reference Zug and Hammerstein2012). They are common and widespread in insects (Werren et al., Reference Werren, Windsor and Guo1995b ), which represent the greatest diversity of all known animal groups on Earth (Rafael et al., Reference Rafael, Melo, de Carvalho, Casari and Constantino2012), equivalent to around 60% of all currently described organisms (Grimaldi & Engel, Reference Grimaldi and Engel2005). They are important for maintenance of ecosystems, as agricultural pests and vectors of human diseases, and useful in medicine and scientific research, besides representing a commercial value food in some cultures (Triplehorn & Johnson, Reference Triplehorn, Johnson and Belmont2005).
Due to the importance of Wolbachia, some researchers have investigated the presence of these bacteria in insects from different locations (Duron et al., Reference Duron, Bouchon, Boutin, Bellamy, Zhou, Engelstädter and Hurst2008; Russell, Reference Russell2012; Russell et al., Reference Russell, Funaro, Giraldo, Goldman-Huertas, Suh, Kronauer, Moreau and Pierce2012). Hilgenboecker et al. (Reference Hilgenboecker, Hammerstein, Schlattmann, Telschow and Werren2008) estimated that over 65% of insect species carry Wolbachia. However, other studies reported that up to 76% (Jeyaprakash & Hoy, Reference Jeyaprakash and Hoy2000) or as few as 20% of insect species are infected with Wolbachia (Werren et al., Reference Werren, Windsor and Guo1995b ). In the first published survey of Wolbachia distribution, Werren et al. (1995b) found over 16% of sampled insect species from Panama were infected with Wolbachia, within several insect orders. In the UK, 22% of insects sampled were infected with Wolbachia, mainly in the Lepidoptera and Hymenoptera (West et al., Reference West, Cook, Werren and Godfray1998). In North America, insect species from 13 different orders were screened for Wolbachia, of which 19.3% were positive. The bacteria have been found in species within several major insect orders: Coleoptera, Diptera, Hymenoptera, Lepidoptera and Orthoptera (Werren & Windsor, Reference Werren and Windsor2000).
Wolbachia detection in Arthropoda has been traditionally performed through standard polymerase chain reaction (PCR) assays targeting the 16S rRNA gene, or protein-coding genes such as the Wolbachia surface protein (wsp) gene and the bacterial cell division gene ftsZ (reviewed in Simões et al. (2011). In contrast, real-time quantitative PCR (qPCR), which possesses high reproducibility, sensitivity and precision of results, has never been used as a tool for Wolbachia screening in Arthropoda.
In Brazil, there are some reports regarding the detection of Wolbachia in limited, specific arthropod groups, but no general surveys of Wolbachia distribution among arthropods have so far been conducted. For example, infection of Wolbachia has been detected in two species of Balloniscus (Crustacea, Oniscidea) (Almerão et al., Reference Almerão, Fagundes, de Araújo, Verne, Grandjean, Bouchon and Araújo2012) and in some species of Diptera in Culicidae (de Albuquerque et al., Reference de Albuquerque, Magalhães and Ayres2011; de Almeida et al., Reference de Almeida, Moura, Cardoso, Winter, Bijovsky and Suesdek2011; Morais et al., Reference Morais, de Almeida, Suesdek and Marrelli2012; Baton et al., Reference Baton, Pacidônio, da Silva Gonçalves and Moreira2013) and in Hymenoptera (Formicidae) (Martins et al., Reference Martins, Souza and Bueno2012). Here, we show the incidence of Wolbachia in different insect orders from the northern and southeastern regions of Brazil using three different markers (16S rRNA, wsp and ftsZ), and the observed incidence corroborates the previously reported widespread nature of this bacterium. We also emphasize the importance of using qPCR for Wolbachia high-throughput screening.
Materials and methods
Insect collection sites
Insects were collected from various field sites spanning the northern and southeastern regions of Brazil, from 2009 to 2012. Samples were obtained from urban, non-urban, forest and forest fragments from Manaus, Careiro da Várzea, Coari and Lábrea in the state of Amazonas; from Belo Horizonte, Belo Vale, Campo Belo and São João da Missões in the state of Minas Gerais; and from Niterói and Rio de Janeiro city in the state of Rio de Janeiro (table 1).
Table 1. Insect collection sites. Insects were collected from different settings: urban, non-urban, forest and forest fragments in northern (Amazonas state) and southeastern (Belo Horizonte and Rio de Janeiro), Brazil (2009–2012).

Insect collection and identification
Insects were manually collected using forceps, nets or traps: HP trap with light attraction (HP Biomédica, Sabará, Minas Gerais, Brazil; Pugedo et al., Reference Pugedo, Barata, França-Silva, Silva and Dias2005), CDC trap+CO2 (John W. Hock Company, Gainesville, Florida, USA) and BG-Sentinel traps (Biogents AG, Regensburg, Germany). Whole insects were individually preserved (to prevent potential cross-contamination) in 96% ethanol and stored at 4°C until identification and DNA extraction. Specimens were identified based on morphology to family level according to Rafael et al. (Reference Rafael, Melo, de Carvalho, Casari and Constantino2012) and Triplehorn & Johnson (Reference Triplehorn, Johnson and Belmont2005). Sand flies were identified to species level through genital morphology according to Galati (Reference Galati, Rangel and Lainson2003) and mosquitoes to species according to Consoli & de Oliveira (Reference Consoli and de Oliveira1994), Faran & Linthicum (Reference Faran and Linthicum1981) and Linthicum (Reference Linthicum1988). Photos were taken for voucher samples with a stereomicroscope (Zeiss Stemi DV4) and digital camera (Canon SX30 IS). Insects that had bristles and spots on the wings, which were important for identification, were not preserved in ethanol but kept in silica.
DNA extraction
Small insects had their bodies homogenized, whereas larger insects were dissected in 1X PBS, to remove ovaries, fat body, thorax and/or abdomen. In the latter case, individual organs were used for DNA extraction.
Crude DNA samples were prepared from individual insects by homogenization in 80 μl ‘squash buffer’ (0.4 mM EDTA, 4 mM Tris, 20 mM NaCl) using a Mini-Beadbeater-16 (BioSpec Products, Inc., Bartlesville, Oklahoma, USA) (modified from Fu et al., Reference Fu, Gavotte, Mercer and Dobson2010). All samples were measured using a NanoDrop (Thermo Scientific Waltham, MA, USA) and diluted to a final concentration of 20 to 50 ng genomic DNA μl−1.
Template and PCR reaction
Insects were screened for the presence of Wolbachia using PCR. Standard PCR was used for the ribosomal 16S rRNA gene with the primers 16S-2 (originally called Wspec; Werren & Windsor, Reference Werren and Windsor2000; Simões et al., Reference Simões, Mialdea, Reiss, Sagot and Charlat2011). Real-time qPCR was performed for the wsp and ftsZ genes using the wsp primers (Moreira et al., Reference Moreira, Iturbe-Ormaetxe, Jeffery, Lu, Pyke, Hedges, Rocha, Hall-Mendelin, Day, Riegler, Hugo, Johnson, Kay, McGraw, van den Hurk, Ryan and O'Neill2009) and newly designed primers to the ftsZ gene, as follows; ftsZqPCR Forward: 5′-GCATTGCAGAGCTTGGACTT-3′ and ftsZqPCR Reverse: 5′-TCTTCTCCTTCTGCCTCTCC-3′. The ftsZqPCR primers were designed using Primer3 (Rozen & Skaletsky, Reference Rozen and Skaletsky2000; Untergasser et al., Reference Untergasser, Cutcutache, Koressaar, Ye, Faircloth, Remm and Rozen2012) to amplify a 271 bp fragment of the ftsZ gene from as broad a spectrum as possible of known sequences from Supergroups A and B, but not C and D, Wolbachia. The specificity of the ftsZqPCR primers to Wolbachia was checked using NCBI Primer-BLAST against the non-redundant database. Control DNA samples were prepared using adult females of the mosquito Aedes aegypti artificially infected with either the wMel (Walker et al., Reference Walker, Johnson, Moreira, Iturbe-Ormaetxe, Frentiu, McMeniman, Leong, Dong, Axford, Kriesner, Lloyd, Ritchie, O'Neill and Hoffmann2011) or wMelPop strains of Wolbachia (McMeniman et al., Reference McMeniman, Lane, Cass, Fong, Sidhu, Wang and O'Neill2009).
Standard PCR had the following components: a final concentration of 0.5X Buffer A and 0.5X Buffer B, 0.13 mM dNTP, 1 μM of each 16S-2 F/R primer, together with 0.3 μl of Elongase (Applied Biosystems®,Grand Island, New York, USA) and a total of 20–50 ng μl−1 of sample DNA, made up with water to a total volume of 25 μl. Amplifications were performed in an automatic thermocycler (Veriti™ Dx Thermal Cycler, Applied Biosystems®,Grand Island, New York, USA) using 35 cycles (30 s 94°C, 30 s 52°C, 1.5 min 68°C) preceded by 5 min at 94°C and followed by a final extension step of 10 min at 68°C. PCR products were visualized on 2% agarose gels stained with Gel Red (diluted 1000×, Biotium, Inc. Hayward, California, USA). qPCR had a final concentration of 1× SYBR® Green PCR Master Mix (Applied Biosystems) and 0.5 μM of each primer (wsp F/R or ftsZqPCR F/R), with a total of 20–50 ng of sample DNA and water to a total volume of 20 μl. The DNA was amplified through 40 cycles (15 s at 95°C and 30 s at 60°C) for the wsp R/F primers, and for 40 cycles (15 s at 95°C, 60 s at 60°C) for the ftsZqPCR F/R primers. All qPCR reactions were carried out in a 96-well microtitre plate (Model 7500, Applied Biosystems). Results were analyzed with the 7500 software v2.0.5, through individual analysis of each amplification curve (compared to the pattern of a positive control) and also their melting curves to check the specificity of the amplification.
In order to confirm the PCR results and therefore, Wolbachia infection status, we sequenced a subset of 61 samples (table 2), that exhibited positive results for only one set of primers. For that, DNA was amplified through conventional PCR under the same conditions as the qPCR (see above). After conventional PCR, the samples were then purified (PCR Purification Kit, Qiagen; Venlo, Limburg, Netherlands), lyophilized and sent for sequencing (Macrogen; Seoul, Korea). As a control, we also sequenced the DNA of A. aegypti artificially infected with the wMelPop (McMeninam et al., Reference McMeniman, Lane, Fong, Voronin, Iturbe-Ormaetxe, Yamada, McGraw and O'Neill2008), using the 16S rRNA, ftsZ and wsp primers. The raw sequencing reads were trimmed and analyzed using the nucleotide-nucleotide BLAST (BLASTN) tool from NCBI and results are shown on table 2.
Table 2. Sequenced insect samples. Insects samples were sequenced for Wolbachia using wsp, 16S rRNA and fstZ primers.

Results
A total of n = 396 insect specimens from 194 species were screened for Wolbachia in 14 orders and 58 families. The largest group belonged to Diptera (n = 191; 48% of all specimens examined) followed by Hemiptera (n = 56; 14%), Hymenoptera (n = 56; 14%) and Coleoptera (n = 34; 9%). The highest number of species belonged to Diptera (n = 65; 34% of all species examined), followed by Hymenoptera (n = 35; 18%), Hemiptera (n = 33; 17%) and Coleoptera (n = 25; 13%) (fig. 1a, b and table 3).
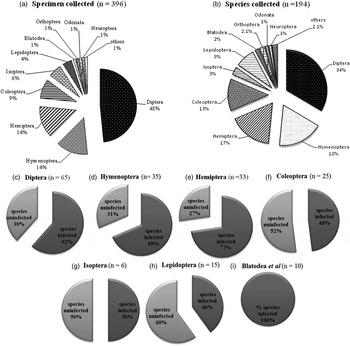
Fig. 1. Diversity and abundance of insects collected, and the proportion of species infected with Wolbachia in each order. In figures C–I: infected (dark grey) and uninfected (light grey) with Wolbachia.
Table 3. Number of insects collected and infected with Wolbachia. Insects were screened for Wolbachia using wsp, 16S rRNA and fstZ primers.

We used three sets of primers to increase the chance of detecting different strains of Wolbachia in our insect samples: 16S rRNA, wsp and ftsZ. We found 28.3% specimens positive for the 16S rRNA marker, 46.2% for wsp and 39.7% for the ftsZ primer (table 3). As expected, the wsp and ftsZ primers were more sensitive in detecting Wolbachia infections than the 16S rRNA primers, which were used for qPCR.
Overall, Wolbachia was found in 10 of the 14 insect orders surveyed, with 232 (58.6%) specimens and 120 (61.9%) species positive. We found 100% species infected with Wolbachia in Orthoptera/Blattodea/Neuroptera/Siphonaptera, 73% in Hemiptera, 69% in Hymenoptera, 62% in Diptera, 50% in Isoptera, 48% in Coleoptera and 40% in Lepidoptera (fig. 1c–i). Wolbachia was not detected in four insect orders: Odonata, Psocoptera, Diplura or Thysanura. This probably reflects the small sample sizes for these insect groups, rather than the absence of Wolbachia, as previous studies have found Wolbachia in the Odonata and Psocoptera (Thipaksorn et al., Reference Thipaksorn, Jamnongluk and Kittayapong2003; Dong et al., Reference Dong, Wang and Zhao2006). Wolbachia were present in 46 families from the 10 PCR-positive orders screened. Orders with the largest number of families infected with Wolbachia were Hemiptera (n = 12; 20.7%), Diptera (12; 21%) and Coleoptera (7; 12.1%) (table 3).
Within Diptera (families Culicidae and Psychodidae) and Hemiptera (Reduviidae), which include several human disease vectors species, we screened 41 species and 19 were positive for Wolbachia (table 3). In Culicidae, we found Wolbachia in four species and two genera. Positive results for Culex quinquefasciatus Say, 1823 and Aedes albopictus (Skuse, 1894) and Culex sp. were expected as their infectious status is widely reported. However, for Mansonia titillans (Walker, Reference Walker, Johnson, Moreira, Iturbe-Ormaetxe, Frentiu, McMeniman, Leong, Dong, Axford, Kriesner, Lloyd, Ritchie, O'Neill and Hoffmann1848), Psorophora cingulata (Fabricius, 1805) and Limatus sp. this is first report of Wolbachia. In Psychodidae, we report here for the first time the presence of Wolbachia in four phlebotomine species: Psychodopygus llanosmartinsi (Fraiha & Ward, 1980), Sciopemyia sordellii (Shannon & Del Ponte, 1927), Psychodopygus davisi (Root, 1934), Trichophoromyia flochi (Abonnenc & Chassignet, 1948), and two genera whose species have not been identified: Evandromyia sp. and Psychodopygus sp. In Reduviidae we did not find Wolbachia in any of screened species of triatominae: Triatoma infestans (Klug, 1934), Panstrongylus megistus (Klug, 1934), Triatoma brasiliensis Neiva, 1911 and Rhodnius prolixus Stål, 1859. These species are exclusively hematophagous, and have been reported with their association with Chagas disease transmission (table 4).
Table 4. Species and genus of hemipterans, Culicidae and phlebotomines collected and screened for Wolbachia. Hemipterans from colony, Culicidae from several localities, and phlebotomines from colony and Amazon.

1 Specimens from CPqRR/Fiocruz colony.
2 Species vectors of disease.
3 Specimens from UFMG (Minas Gerais) colony.
Sequencing a subset of samples allowed us to confirm the majority of samples that showed positivity with the PCR analysis. From a total of 61 DNA samples, 46 returned sequences belonging to Wolbachia (table 2).
Discussion
We studied the incidence of Wolbachia in insects collected from northern and southern parts of Brazil. Most of the insects collected belong to Coleoptera, Diptera, Hemiptera and Hymenoptera. Although we used light and CO2 traps, as well as manual sampling to collect insects near or within urban areas, targeting a great diversity of insect groups, most of the insects sampled were Diptera, Heteroptera, Hymenoptera and Coleoptera. This is because these orders are large and well-diversified, making it easier to collect representatives in different habitats. The higher prevalence of Wolbachia in Diptera was expected, since many species in this order have previously been reported to be infected with the endosymbiont, and we collected more specimens and species from this order, so that we would be more likely to detect rare infections (Duron et al., Reference Duron, Bouchon, Boutin, Bellamy, Zhou, Engelstädter and Hurst2008; Hilgenboecker et al., Reference Hilgenboecker, Hammerstein, Schlattmann, Telschow and Werren2008; Zug & Hammerstein, Reference Zug and Hammerstein2012). In dipteran insects, especially mosquitoes (Hertig & Wolbach, Reference Hertig and Wolbach1924) and drosophilids, Wolbachia is commonly found (Boyle et al., Reference Boyle, O'Neill, Robertson and Karr1993; Braig et al., Reference Braig, Guzman, Tesh and O'Neill1994). Furthermore, many other insect groups are known to carry Wolbachia: e.g., leafhoppers, thrips and whiteflies (Nirgianaki et al., Reference Nirgianaki, Banks, Frohlich, Veneti, Braig, Miller, Bedford, Markham, Savakis and Bourtzis2003), termites (Bandi et al., Reference Bandi, Sironi, Nalepa, Corona and Sacchi1997; Lo et al., Reference Lo, Casiraghi, Salati, Bazzocchi and Bandi2002; Bordenstein & Rosengaus, Reference Bordenstein and Rosengaus2005), beetles (Werren & Windsor, Reference Werren and Windsor2000; Nirgianaki et al., Reference Nirgianaki, Banks, Frohlich, Veneti, Braig, Miller, Bedford, Markham, Savakis and Bourtzis2003), odonates (dragonflies and damselflies) (Thipaksorn et al., Reference Thipaksorn, Jamnongluk and Kittayapong2003) and crickets (Kamoda et al., Reference Kamoda, Masui, Ishikawa and Sasaki2000). Although in our collections, Hemiptera and Hymenoptera had fewer species and specimens collected compared to Diptera, Wolbachia had a higher incidence.
Heteroptera, known as true bugs, is one of the most diverse groups of insects with incomplete metamorphosis. Wolbachia infection was previously reported in this group (Kikuchi & Fukatsu, Reference Kikuchi and Fukatsu2003) and here we observed a 28.6% frequency of infection distributed in eight different families (Gerridae, Corixidae, Cydnidae, Berytidae, Pyrrhocoridae, Rhopalidae, Pentatomidae and Coreidae), six of them previously reported by Kikuchi & Fukatsu (Reference Kikuchi and Fukatsu2003). In many groups of Heteroptera, the removal of the endosymbionts can result in stunted growth and/or mortality of the nymphs, suggesting a major role for Wolbachia in this host association (Fukatsu & Hosokawa, Reference Fukatsu and Hosokawa2002).
Wolbachia also influence reproductive patterns in social Hymenoptera. Studies on ants in Indonesia showed that Wolbachia was common, with 50% of the species infected (Wenseleers et al., Reference Wenseleers, Ito, Van Borm, Huybrechts, Volckaert and Billen1998). In our study, from 13 species of ants screened, nine were infected with Wolbachia, representing an incidence of infection greater than 69%. Wolbachia infection has been reported to cause parthenogenesis in some families of Coleoptera (Werren et al., Reference Werren, Zhang and Guo1995a ; Rodriguero et al., Reference Rodriguero, Confalonieri, Guedes and Lanteri2010). Furthermore, evidence of horizontal transfer of Wolbachia was also found in Curculionidae, Chrysomelidae and Tenebrionidae (Rodriguero et al., Reference Rodriguero, Confalonieri, Guedes and Lanteri2010). We collected 19 species of beetles from these and others families. Wolbachia was present in 12 species: Cantharidae (n = 1), Chrysomelidae (n = 3), Curculionidae (n = 2), Haliplidae (n = 1), Cerambicidae (n = 1), Anobiidae (n = 1), Brentidae (n = 1) and two other species. Based on 16S rRNA and wsp sequence detection, Wolbachia had already been reported in siphonapteran hosts (Jeyaprakash & Hoy, Reference Jeyaprakash and Hoy2000; Gorham et al., Reference Gorham, Fang and Durden2003; Dittmar & Whiting, Reference Dittmar and Whiting2004) and in this study we collected a flea [Ctenocephalides canis Curtis (Siphonaptera, Pulicidae)] from a domestic dog that was also positive for Wolbachia. According to Dittmar & Whiting (Reference Dittmar and Whiting2004), the discovery of symbiotic bacteria in wild populations of Siphonaptera suggests a potentially widespread association with fleas. Although we collected only two specimens of the same species, one specimen was positive.
In the present study, the overall incidence of Wolbachia among species was similar to that reported by Hilgenboecker et al. (Reference Hilgenboecker, Hammerstein, Schlattmann, Telschow and Werren2008) who estimated that the percentage of infected Wolbachia species is approximately 66%, when rarely infected species are included. Most of the species that we screened were based on one or only a few individuals. Within each species from the same population, we found that 40 to 100% specimens were infected with Wolbachia (i.e., the intra-specific prevalence of Wolbachia varied from low to high frequency). This could be because the levels of infection within a host population may depend on the age of the endosymbiont–host association (i.e., whether there has been sufficient time for Wolbachia to invade the host population) and how Wolbachia manipulates the reproduction of their hosts (Hurst & Jiggins, Reference Hurst and Jiggins2000).
Wolbachia is naturally present in many genera of mosquitoes, including Aedes, Culex, Mansonia and Coquillettidia (Kittayapong et al., Reference Kittayapong, Baisley, Baimai and O'Neill2000; Ricci et al., Reference Ricci, Cancrini, Gabrielli, D'Amelio and Favia2002; Dean & Dobson, Reference Dean and Dobson2004) and recently it has been reported in Anopheles gambiae (Baldini et al., Reference Baldini, Segata, Pompon, Marcenac, Robert Shaw, Dabiré, Diabaté, Levashina and Catteruccia2014). Our survey also revealed the presence of Wolbachia in a number of other potential vectors of human pathogens. Wolbachia has previously been found in the gonads and salivary glands of Rhodnius pallescens Barber, 1932, which is considered the most important vector of Trypanosoma cruzi and Trypanosoma rangeli in the Neotropics (Espino et al., Reference Espino, Gómez, González, do Santos, Solano, Sousa, Moreno, Windsor, Ying, Vilchez and Osuna2009), but the role of this endosymbiont in the relationship between the insect and parasite is not yet known. In Brazil, there are several kissing bug species, which are important vectors of Chagas disease, such as T. infestans, T. brasiliensis, R. prolixus and P. megistus (Costa & Lorenzo, Reference Costa and Lorenzo2009), but there are no reports about the presence of Wolbachia in these insects. Although the wsp marker detected Wolbachia in five specimens of P. megistus and T. brasiliensis, while the ftsZ primers detected the bacterium in two specimens of R. prolixus and one P. megistus, the infection was not confirmed by sequencing (table 2), as the blasted sequences had no hits to Wolbachia. It is important to emphasize that these particular samples were derived from the laboratory. Broader screening of field specimens should be envisaged, increasing the chance of Wolbachia detection.
Wolbachia has also been reported in the Phlebotominae (Diptera: Psychodidae) both in New (Ono et al., Reference Ono, Braig, Munstermann, Ferro and O'Neill2001; Azpurua et al., Reference Azpurua, De La Cruz, Valderama and Windsor2010) and Old World species (Zhou et al., Reference Zhou, Rousset and O'Neill1998). Phlebotomines are vectors of several viral, bacterial and protozoal diseases of humans and other animals, but there are few studies on the presence of Wolbachia in sand flies (Cui et al., Reference Cui, Chang, Strickman and Rowton1999; Ono et al., Reference Ono, Braig, Munstermann, Ferro and O'Neill2001; Benlarbi & Ready, Reference Benlarbi and Ready2003; Matsumoto et al., Reference Matsumoto, Izri, Dumon, Raoult and Parola2008; Azpurua et al., Reference Azpurua, De La Cruz, Valderama and Windsor2010; de Sousa et al., Reference de Sousa, da Silva, Alencar, Baton, Naveca and Shimabukuro2013) and about the biological relationship of the endosymbiont with the host (Kassem et al., Reference Kassem, Hassan, Abdel-Hamid, Osman, El Khalab and Madkour2003; Kassem & Osman Reference Kassem and Osman2007). In Iran, a new strain of Wolbachia was recently found in Phlebotomus perfiliewi transcaucasicus Perfil'ev, 1937 (Parvizi et al., Reference Parvizi, Fardid and Soleimani2013), increasing the list of phlebotomines known to be infected with this endosymbiont. Further studies should explore the potential for Wolbachia to be used as a biological control agent for Leishmania vectors. Here, we collected 21 sand fly species (20 wild species from Amazonas and one from a colony), and Wolbachia was found only in wild species. In six wild species, the bacterium was found using both wsp and ftsZ primers. Only in a single wild species of the genus Evandromyia was Wolbachia detected by all three markers.
Conclusions
Due to the high diversity amongst different Wolbachia strains, it is difficult to detect a wide range of strains using one set of universal primers. Currently, new strains of Wolbachia in different host species have been found, mainly due to the use of a combination of primers to improve detection of this bacterium (Lo et al., Reference Lo, Casiraghi, Salati, Bazzocchi and Bandi2002). Here, we used three different primer sets and two PCR methods to enhance the detection of Wolbachia in an extensive collection of insects. According to Simões et al. (Reference Simões, Mialdea, Reiss, Sagot and Charlat2011), the 16S rRNA primers are sensitive to detect a broad-spectrum of Wolbachia. However, these primers do not detect all Wolbachia strains. It was clear in our results that the primers used for real-time qPCR (wsp and ftsZ) showed a higher number of positive samples than conventional PCR (using the 16S rRNA primer set), which can be explained by the higher sensitivity provided by qPCR.
In summary, one should take into account the difficulty of designing primers covering all existing groups of Wolbachia, but on the other hand be cautious of using a single marker, such as wsp or ftsZ, as this could potentially underestimate Wolbachia prevalence in a given sample. Finally, we recommend the use of real-time qPCR because it is the most sensitive and fastest method to detect Wolbachia in a wide variety of arthropod samples.
Acknowledgements
We would like to thank Wanderli Tadei, Alessandra Guarneri, Liléia Diotaiuti and Rafael Freitas for providing specimens and Edelberto Dias for lending us traps. Also, Taís Souza for helping with sequencing analysis. The authors thank the Program for Technological Development in Tools for Health-PDTIS-FIOCRUZ for using its facilities. Financial support was provided by INCT-EM, FAPEMIG and CNPq. LAM is a CNPq fellow.








