Introduction
The Southern Ocean began to cool approximately 40 million years ago (m.y.a.), reaching the current temperature range of +2°C to -1.8°C approximately 25 m.y.a. (Clark et al. Reference Clark, Clarke, Cockell, Convey, Detrich, Fraser, Johnston, Methe, Murray, Peck, Römisch and Rogers2004). The formation of ice crystals in the water would normally be fatal for living organisms, due to the physical destruction of membranes and the resulting chemical damage from processes such as osmotic shock. Antarctic organisms have developed a variety of molecular strategies to thrive in these low-temperature environments, such as increasing molecular flexibility to modulate the kinetics of key enzymes and developing biological membranes that are more fluid through the accumulation of polyunsaturated fatty acyl chains (Morgan-Kiss et al. Reference Morgan-Kiss, Priscu, Pocock, Gudynaite-Savitch and Huner2006). Another adaptive strategy involves the use of ice-binding proteins (IBPs), which include antifreeze proteins (AFPs), to combat the destructive effects of ice crystals. IBPs can inhibit ice growth by binding to specific ice planes and lowering the freezing point to below the melting point of the ice. The difference between the freezing and melting temperatures is called thermal hysteresis (TH) and is used as a measure of antifreeze activity (Scotter et al. Reference Scotter, Marshall, Graham, Gilbert, Garnham and Davies2006).
Initially identified in Antarctic marine fishes, AFPs protect their hosts from freezing by binding to, and preventing the growth of, seed ice crystals (DeVries & Wohlschl Reference DeVries and Wohlschlag1969). In fishes, four types of AFPs have been characterized: types I, II and III, and antifreeze glycoproteins (Fletcher et al. Reference Fletcher, Hew and Davies2001); interestingly, these AFPs are apparently unrelated. Type II AFPs, which are derived from C-type lectin genes, are the only type that have been found in three phylogenetically disparate fish species: the Atlantic herring (Clupeiformes), the smelt (Salmoniformes) and the sea raven (Scorpaenoformes). This finding suggests that type II AFPs have evolved through the duplication and divergence of the C-type lectin genes at least three times (Ewart et al. Reference Ewart, Rubinsky and Fletcher1992). Subsequently, AFPs were found in other freeze-tolerant organisms, such as plants (Sidebottom et al. Reference Sidebottom, Buckley, Pudney, Twigg, Jarman, Holt, Telford, McArthur, Worrall, Hubbard and Lillford2000), fungi (Kondo et al. Reference Kondo, Hanada, Sugimoto, Hoshino, Garnham, Davies and Tsuda2012) and bacteria (Raymond et al. Reference Raymond, Christner and Schuster2008, Do et al. Reference Do, Lee, Lee and Kim2012). The bacterial AFPs stop the growth of ice crystals through ice re-crystallization inhibition (IRI) in frozen tissues, rather than preventing the freezing of tissues (Gilbert et al. Reference Gilbert, Hill, Dodd and Laybourn-Parry2004). Gilbert et al. (Reference Gilbert, Hill, Dodd and Laybourn-Parry2004) isolated 11 bacterial strains from Antarctica with IRI activity. Among these, Marinomonas primoryensis Romanenko et al. produces a hyperactive, Ca2+-dependent AFP, which lowers the freezing point of water by 2°C (Gilbert et al. Reference Gilbert, Hill, Dodd and Laybourn-Parry2004). This mode of action resembles that of insect AFPs (Duman Reference Duman2001); ice crystals formed in the presence of this AFP do not have distinct facets and are typically round in shape.
Over the past decade, IBPs/AFPs have proven to be potent agents in the cryopreservation of food and pharmaceutical materials. For example, the AFP from carrots (Daucus carota L.) has been shown to improve the leavening of frozen dough, and to help maintain loaf volume and dough softness during frozen storage (Yeh et al. Reference Yeh, Kao and Peng2009). Additionally, the type I AFP produced in Lactococcus lactis (Lister) improves the fermentation capacity of frozen dough and the conservation of frozen meat (Yeh et al. Reference Yeh, Kao and Peng2009).
The mechanisms of molecular adaptation to low-temperature environments remain poorly understood, largely due to the limited amount of available molecular data. To obtain sufficient sample data necessary to identifying the molecular adaptation in psychrophiles, the entire macronuclear genome from the Antarctic ciliate Euplotes focardii Valbonesi & Luporini was sequenced recently. Using this approach, we expected to discover new genes that may be responsible for cold-adaptation, including genes encoding IBPs.
Euplotes focardii was isolated from the coastal seawater near Terra Nova Bay and is endemic to the Antarctic continent (Valbonesi & Luporini Reference Valbonesi and Luporini1993). It has been maintained in laboratory culture since its first isolation. Its psychrophilic phenotypes include optimal survival and multiplication rates at temperatures between 4–5°C (Valbonesi & Luporini Reference Valbonesi and Luporini1993), lack of a transcriptional response of the Hsp70 genes to thermal stress (La Terza et al. Reference La Terza, Papa, Miceli and Luporini2001), and modifications in the primary structure of the tubulin (Marziale et al. Reference Marziale, Pucciarelli, Ballarini, Melki, Uzun, Ilyin, Detrich and Miceli2008, Pucciarelli et al. Reference Pucciarelli, La Terza, Ballarini, Barchetta, Yu, Marziale, Passini, Methé, Detrich and Miceli2009, Chiappori et al. Reference Chiappori, Pucciarelli, Merelli, Ballarini, Miceli and Milanesi2012) and ribosomal stalk proteins (Pucciarelli et al. Reference Pucciarelli, Marziale, di Giuseppe, Barchetta and Miceli2005). As in all ciliates, E. focardii possesses two nuclei: a diploid micronucleus involved only in sexual exchange and a polyploid macronucleus responsible for all gene expression, which is renewed after each phenomenon of sexuality (Hoffman et al. Reference Hoffman, Anderson, Dubois and Prescott1995). In the latter, the genome is organized in small linear chromosomes (nanochromosomes). Each nanochromosome contains a single genetic unit that is flanked by regulatory regions and capped by telomeres (Hoffman et al. Reference Hoffman, Anderson, Dubois and Prescott1995). Among the assembled nanochromosomes characterized by regular Euplotes telomeres, DNA sequences that lack telomeres and thus do not belong to the E. focardii nanochromosomes have been identified. We report the identification and sequence analysis of two IBP genes obtained from the E. focardii genome.
Materials and methods
Materials
Taq polymerase, DNA modifying enzymes and restriction enzymes were purchased from Fermentas (Milan, Italy). Oligonucleotides were synthesized by Sigma/Genosys (Milan, Italy). All routine chemicals were of analytical grade and supplied by Sigma Aldrich (Milan, Italy).
Cell strains and growth conditions
Cell cultures of E. focardii strain TN1 (Valbonesi & Luporini Reference Valbonesi and Luporini1993) were used for this study. They were isolated from coastal sediment and seawater samples collected in Terra Nova Bay, Ross Sea (temperature -1.8°C, salinity 35‰, pH 8.1–8.2). The cultures were grown at 4°C and fed on the green algae Dunaliella tertiolecta Butcher.
Isolation of Euplotes focardii macronuclear DNA and rapid amplification of telomeric ends
The macronuclear DNA of E. focardii was purified as described by Miceli et al. (Reference Miceli, La Terza and Melli1989). Organization in small nanochromosomes facilitates the characterization of the 5’- and 3’-untranslated region sequences using the rapid amplification of telomeric ends-PCR (RATE-PCR) technique (Pucciarelli et al. Reference Pucciarelli, Marziale, di Giuseppe, Barchetta and Miceli2005, Marziale et al. Reference Marziale, Pucciarelli, Ballarini, Melki, Uzun, Ilyin, Detrich and Miceli2008). To determine whether the genes encoding the AFPs were present in the E. focardii genome, we used degenerate forward and reverse primers designed against the conserved amino acid sequences (Table I).
Table I Degenerate forward and reverse primers designed against the conserved amino acid (AA) sequences, used to determine whether the genes encoding the antifreeze proteins were present in the E. focardii genome.
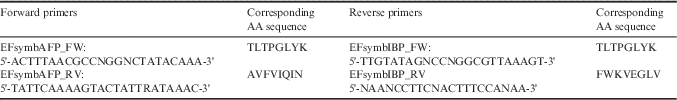
The PCR reactions were started with only one of the listed primers. After ten cycles, the telomeric oligonucleotide 5’-(C4A4)4-3’ was added to complete the strategy: 1 minute at 94°C, 1 minute at 55°C and 1 minute at 72°C for 35 cycles. Amplified products were cloned into the pTZ57R/T vector following the procedure recommended by the manufacturer (Fermentas, Milan, Italy). The recombinant vectors were sequenced by BMR Genomics (Padua, Italy).
Colony blotting
Colony blotting was performed as previously described by Marziale et al. (Reference Marziale, Pucciarelli, Ballarini, Melki, Uzun, Ilyin, Detrich and Miceli2008). Probes corresponding to the sequence of the AFP genes from the E. focardii symbiont were used, following the procedure described by Yu et al. (Reference Yu, Barchetta, Pucciarelli, La Terza and Miceli2012).
Sequence analysis, alignment and phylogenetic tree construction
Sequences were obtained from the analysed reads and assembled contigs of the E. focardii macronuclear genome sequencing performed in collaboration with Dr Vadim Gladishev’s research group (Harvard Medical School, USA).
For the tBLASTn analysis, we used the Fragilariopsis cylindrus (Grunow) Krieger AFP sequences (GenBank: ACT21449.1, ACT21447.1, ACT21445.1, ACT21443.1, ACT21448.1, ACT21446.1, ACT21444.1, ABT17158.1, ACX36856.1, ACX36854.1, ACX36852.1, ACX36855.1 and ACX36853.1), a dominant species within the polar sea ice assemblages (Bayer-Giraldi et al. Reference Bayer-Giraldi, Uhlig, John, Mock and Valentin2010), to query the genome of E. focardii.
To verify the tandem organization of the two IBP gene sequences on the bacterial chromosome, we performed PCR using the forward primer 5’-ACAACAACACCAGGGCGAAAAG-3’ and the reverse primer 5’-TTCGTTGTAAGCGATTAATAAATC-3’. We used macronuclear DNA isolated from unperturbed E. focardii cells or from cells treated with antibiotics to remove the symbiotic bacteria (Vannini et al. Reference Vannini, Schena, Verni and Rosati2004). DNA from the cells treated with antibiotics was used as a control. The expected band was of c. 750 bp. Furthermore, to exclude the possibility that the IBP sequences were derived from associated bacteria or endosymbionts of the algae in the culture medium rather than from the Euplotes, we performed a PCR reaction using DNA purified from E. focardii or Dunaliella cells.
Sequence similarity and conserved domain analysis were performed using the Basic Local Alignment Search Tool (BLAST) program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using the default parameters. Multiple sequence alignments were performed using the ClustalW2 tool (http://www.ebi.ac.uk/Tools/msa/clustalw2/) or the PRALINE tool (http://www.ibi.vu.nl/programs/pralinewww/) using the default parameters. Percentage similarities were estimated using the ClustalW2 tool. The evolutionary history was inferred using the maximum parsimony (MP) method. The MP tree was obtained using the Subtree-Pruning-Regrafting (SPR) algorithm with search level 5 in which the initial trees were obtained by the random addition of sequences (ten replicates). The analysis involved 22 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 118 positions in the final dataset. Evolutionary analyses were conducted in MEGA 5 (Tamura et al. Reference Tamura, Peterson, Peterson, Stecher, Nei and Kumar2011).
The sequences were obtained from the National Center for Biotechnology Information (NCBI) database: Sphaerochaeta globus Abt et al. (strain Buddy) YP_004248731, Navicula glaciei Van Heurck AAZ76251, Fragilariopsis cylindrus Grunow, Willi Krieg. CN212299, Fragilariopsis curta (Van Heurck) Hustedt ACT99634, Chlamydomonas sp. EU190445, Typhula ishikariensis S.Imai BAD02897, Flammulina populicola Redhead & Petersen ACL27144, Lentinulaedodes edodes (Berk.) Pegler ACL27145, Glaciozyma antarctica Thomas-Hall & Boekhout ACX31168, Psychromonas ingrahamii Auman et al. ZP_01349469, Colwellia sp. DQ788793, Deschampsia antarctica E.Desv. ACN38296, Lolium perenne L. ACN38303, Marivirga tractuosa (Lewin) (strain DSM4126) YP_004052221, Cytophaga hutchinsonii Winogradsky (strain ATCC33406) YP_676864, Chaetoceros neogracile VanLandingham ACU09498, Nitzschia stellata Manguin AEY75833, Psychroflexus torquis Bowman et al. (strain ATCC700755) YP_006867144, Stigmatella aurantiaca Berkeley & Curtis (strain DW4/3-1) ZP_01462925.1, an uncultured bacterium (EKD52074.1), and Flavobacteriaceae bacterium Bernardet et al. (strain 3519-10) gi_255534643.
Modelling of tertiary structures and putative cleavage site prediction
The 3D structures of EFsymbAFP and EFsymbIBP were modelled, with a homology modelling method, on the structure of the snow mould fungus T. ishikariensis AFP (PDB: 3VN3_A) (Kondo et al. Reference Kondo, Hanada, Sugimoto, Hoshino, Garnham, Davies and Tsuda2012) and the IBP from Arctic yeast Leucosporidium (sp. AY30, PDB: 3UYU_A; LeIBP) (Lee et al. Reference Lee, Park, Do, Park, Moh, Chi and Kim2012) using the routine multiple template model from MODELLER 9v11 (Eswar et al. Reference Eswar, Webb, Marti-Renom, Madhusudhan, Eramian, Shen, Pieper and Sali2006). EFsymbAFP displays a sequence identity of 35% and 34% compared to the fungal AFP and the Arctic yeast LeIBP, respectively; whereas EFsymbIBP displays 33% and 28% of sequence identity, respectively. Models were obtained from the alignment with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo) using default parameters. The resulting models were visualized using the PyMol software. The putative cleavage site of the signal peptide was predicted using the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/).
Results
Identification and analysis of the ice-binding protein sequences
As a first approach to characterizing the IBPs in E. focardii, the entire macronuclear genome was explored using the AFP sequences from the diatom Fragilariopsis cylindrus as query sequences. A 3221 bp contig (KC691985) differed from the typical Euplotes nanochromosomes, i.e. it lacked the telomeric sequences at the ends of the nanochromosomes. Computer-assisted analysis revealed two open-reading frames (ORFs; shaded in grey in Fig. 1). The first ORF encodes a polypeptide of 251 amino acids, similar to the AFP from the DW4/3-1 strain of S. aurantiaca (NC_014623.1, max. identity: 54%, coverage: 91%, e-value: 1e-179), a bacterium isolated from the lower glacier of Victoria Valley. The second ORF encodes a polypeptide of 540 amino acids that is similar to the IBP from the 3519-10 strain of F. bacterium (EU694412, max. identity: 62%, coverage: 81%, e-value: 4e-75), a bacterium isolated from the glacial ice of Lake Vostok at a depth of more than 3000 m. To exclude the possibility that these sequences were derived from associated bacteria or endosymbionts of the algae in the culture medium, a PCR reaction using DNA purified from E. focardii or Dunaliella cells was performed. Only the sample containing E. focardii DNA gave the expected band of 750 bp (Supplemental Fig. S1 found at http://dx.doi.org/10.1017/S0954102014000017). Considering the possibility that these sequences were derived from a symbiotic bacterium of ciliate E. focardii, we named the two sequences EFsymbAFP and EFsymbIBP.

Fig. 1 Partial nucleotide sequence of the bacterial symbiont contig (3211 bp; contig 03294) identified in the E. focardii genome. The two open reading frames encoding for the ice-binding protein and antifreeze protein, respectively, are shaded in grey.
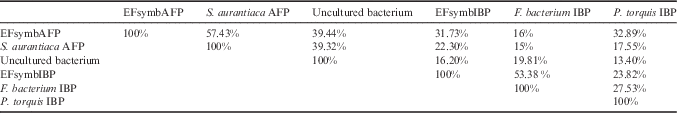
EFsymbAFP is 57.43% identical to the AFP from S. aurantiaca, and 39.44% identical to the AFP from an uncultured bacterium obtained from a metagenomic environmental sample situated at 1645 m above sea level (Rifle, CO, USA) (Table II). The low percentages are due to the different lengths of the two sequences (Fig. 2). All these proteins contain a predicted signal peptide (shaded in grey in Fig. 2). The N-terminal half of the S. aurantiaca and uncultured bacterium AFPs is completely absent in EFsymbAFP. However, EFsymbAFP overlaps fairly well with the N-terminus of a number of IBPs, such as that from Acanthamoeba castellanii strain Neff sh90004976 (XP_004344590).

Fig. 2 Sequence comparison of EFsymbAFP with the antifreeze proteins (AFPs) from S. aurantiaca (ZP_01462925.1) and an uncultured bacterium (EKD52074.1), using the ClustalW2 multiple sequence alignment tool. The predicted secretory signal at the N-terminus is shaded.
Table II Percentage identities of ice-binding protein sequences from the E. focardii symbiont and from other cold-adapted bacteria.
Percentage identities were obtained from the ClustalW2 tool (http://www.ebi.ac.uk/Tools/msa/clustalw2/). AFP=antifreeze protein, IBP=ice-binding protein.
EFsymbIBP is 53.38% identical to the IBP from F. bacterium, and 23.82% identical to that from P. torquis (YP_006867144) a psychrophilic species from Antarctic sea ice (Bowman et al. Reference Bowman, McCammon, Lewis, Skerratt, Brown, Nichols and McMeekin1998) (Table II). The less conserved region is the predicted signal peptide (shaded in grey in Fig. 3).

Fig. 3 Sequence comparison of EFsymbIBP with the ice-binding proteins (IBPs) from F. bacterium (gi_255534643) and P. torquis (strain ATCC700755, YP_006867144), using the ClustalW2 multiple sequence alignment tool. The predicted secretory signal at the N-terminus is shaded.
Both EFsymbAFP and EFsymbIBP, as well as many other AFPs, possess the DUF3494 family domain of unknown function, which is duplicated in EFsymbIBP. This characteristic suggests that the DUF3494 family domain may be a fingerprint for IBPs. The multiple sequence alignments of EFsymbAFP and EFsymbIBP indicate that these sequences maintain similar physical-chemical properties with respect to those from other polar and non-polar organisms, although the amino acid composition is not identical (see Supplemental Figs S2 and S3 found at http://dx.doi.org/10.1017/S0954102014000017).
Further analysis of EFsymbIBP revealed that the N-terminal and C-terminal halves of the protein are nearly 42% similar, suggesting that the protein is the result of gene duplication. This hypothesis is supported by the model of the tertiary structure. Both the N- and C-terminal domains of EFsymbIBP displayed predicted 3D structures that were similar to the T. ishikariensis and Leucosporidium AFPs (Fig. 4). The main structural element is an irregular β-helix, composed of six, non-sequentially arranged loops that lie alongside an α-helix. The ice-binding site (IBS) lies on the flattest β-helical surface of the protein, defined as the b-face (indicated in grey in Fig. 4). The b-face of the Arctic and Antarctic yeast AFPs are composed of six β-sheets (Fig. 4), whereas those from the bacterial symbiont appear to be composed of four (EFsymbAFP) or five (EFsymbIBP, both N- and C-terminal domains) β-sheets (Fig. 4).

Fig. 4 Molecular models of EFsymbAFP and EFsymbIBP. The 3D structures were modelled using the routine multiple template model from MODELLER 9v11 (Eswar et al. Reference Eswar, Webb, Marti-Renom, Madhusudhan, Eramian, Shen, Pieper and Sali2006). a. Antifreeze protein (AFP) structure from the snow mould fungus T. ishikariensis (PDB: 3VN3_A) (Kondo et al. Reference Kondo, Hanada, Sugimoto, Hoshino, Garnham, Davies and Tsuda2012). b. AFP structure from the Arctic yeast Leucosporidium sp. AY30 (PDB: 3UYU_A) (Lee et al. Reference Lee, Park, Do, Park, Moh, Chi and Kim2012). c. EFsymbAFP. d. EFsymbIBP N-terminal domain. e. EFsymbIBP C-terminal domain. f. Sequence alignment of the ice-binding sites (IBSs) from the T. ishikariensis AFP, EFsymbAFP and EFsymbIBP (N- and C-terminal domains). Hydrophobic residues are in yellow and residues putatively involved in hydrogen bonds are in red. The residues shaded in grey represent the IBS of the T. ishikariensis AFP.
Through sequence and structural homology, we identified the IBSs in EFsymbAFP and EFsymbIBP. As reported for the homologues from bacteria, diatoms and fungi, the IBSs are poorly conserved at the sequence level (Fig. 4f). However, they maintain a high content of hydrophobic residues and repetitions of residues that may form hydrogen bonds, including serine, threonine and asparagine, suggesting a mechanism of ice crystal binding by the ‘anchored clathrate’ mode of action.
Genomic organization of the antifreeze proteins
To determine whether the AFP genes of the S. aurantiaca strain DW4/3-1 and the F. bacterium strain 3519-10 were organized in tandem in their respective chromosomes, as with those found in the E. focardii symbiont, the entire genome from both species was analysed (GenBank database: NC_014623 and NC_013062, respectively). Both bacterial chromosomes contained only one copy of the genes encoding the AFPs, suggesting that either a gene duplication or a horizontal gene transfer (HGT) event occurred in the E. focardii symbiont. If the former hypothesis were true, the two bacterial sequences must share an amino acid sequence identity higher than that shared with either the S. aurantiaca or F. bacterium AFPs. The two symbiont sequences are only 31.73% identical (Table II), a percentage that is lower than the shared sequence identities with either the S. aurantiaca or F. bacterium AFP. Therefore, the low identity of the two sequences excludes the gene duplication hypothesis. The construction of a phylogenetic tree (Fig. 5) revealed that the two sequences are more closely related to their orthologs from S. aurantiaca and F. bacterium than to each other. Therefore, the tandem organization of the two AFP gene sequences in the E. focardii symbiont was most probably derived from an HGT event between bacterial strains.

Fig. 5 Maximum parsimony phylogenetic tree based on the amino acid sequences of antifreeze (AFP) and ice-binding proteins (IBP) from bacteria, diatoms, fungi, plants and protists. The most parsimonious tree with length = 1016 is shown. For the parsimony-informative sites, the consistency index is 0.681000, the retention index is 0.495253 and the composite index is 0.337267 (the composite index for all sites is 0.339755). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches.
To exclude the possibility that the tandem organization of the two genes was the consequence of a misassembly of the genomic reads, the assembly of the reads that overlap on the two genes were checked manually. By BLASTing a fragment of 700 bp from the E. focardii genomic contigs, which included the C-terminal domain of the AFP and the N-terminal domain of the IBP, nearly 100 reads that covered the entire fragment of 700 bp were found (Fig. 6a). A PCR reaction using DNA extracted from untreated cells or from cells treated with antibiotics, only produced a reaction with the untreated cells (Fig. 6b), confirming that the two IBP sequences are of bacterial origin and are organized in tandem on the bacterial chromosome.

Fig. 6 Tandem organization of the two ice-binding protein gene sequences. a. Schematic representation of the 700 bp fragment of the E. focardii genomic contig (contig 03294) containing the C-terminal domain of EFsymbAFP and the N-terminal domain of EFsymbIBP; this contig was used to search for the corresponding genomic reads (indicated in pink). b. Agarose gel of the PCR reactions performed using DNA extracted from untreated cells (lane 2) or cells treated with antibiotics (lane 3). The arrow indicates the PCR product obtained in the reaction from the untreated DNA sample.
The HGT may have occurred between the bacteria and E. focardii. If this had occurred, the sequences would be found in the E. focardii genome within the typical nanochromosome organization, i.e. short molecules containing a single ORF protected at the extremities by telomeres composed of the nucleotide repetition C4A4. Therefore, a RATE-PCR was performed using degenerated primers (according to the codon usage of E. focardii) that corresponded to conserved motives of the coding region of the AFPs and primers that bind the telomeric sequences. The PCR products were analysed on an agarose gel (data not shown). In the reaction performed with forward primers (FW and C4A4), no PCR products were detected. However, in the reaction performed with reverse primers (RV and C4A4), a smear was obtained which was subsequently subcloned. The resulting colonies were screened using a probe that corresponded to the AFP coding region. However, this screening method did not identify any positive clones. Sequencing randomly chosen clones revealed that the PCR products were derived from non-specific binding of the primers. These results confirm that these bacterial sequences are not represented in the E. focardii genome, unless they are contained in exceptionally long nanochromosomes. This finding, together with the inability to amplify the genes following antibiotic treatment, corroborates the hypothesis that the AFP and IBP genes are of bacterial origin and excludes the possibility of HGT moving into the E. focardii genome.
Phylogenetic analysis
To assess the phylogenetic relationships between the IBPs encoded by the putative E. focardii symbiont and similar proteins from other organisms, a phylogenetic tree was constructed using the coding sequences of both EFsymbAFP and EFsymbIBP (Fig. 5), as well as the AFPs/IBPs from different species of bacteria, diatoms, fungi and plants.
EFsymbAFP, the AFPs from S. aurantiaca and an uncultured bacterium clustered with the AFP from the Antarctic yeast G. antarctica (Fig. 5). This cluster appears to be the sister group of the clade containing the AFPs/IBPs from all other bacteria, including the Arctic Psychromonas and the Antarctic Colwella, and the clade that includes other Basidiomycota species and diatoms. In contrast, EFsymbIBP and the IBP from the Antarctic bacteria F. bacterium and P. torquis clustered with those from Chlamydomonas and plants. These latter sequences appear to derive from the P. torquis IBP, most probably as a consequence of HGT.
The lineage of AFPs/IBPs forms two separate clades, those from bacteria, fungi and diatoms, and those from Chlamydomonas and plants, suggesting that these proteins followed different evolutionary histories.
Discussion
We report the identification and sequence analysis of two bacterial IBPs, EFsymbAFP and EFsymbIBP. The corresponding bacterial contig was found among the genomic sequences of the Antarctic ciliate E. focardii, suggesting that these sequences belong to an unidentified bacterial symbiont of the ciliate. We excluded the possibility that these sequences are derived from associated bacteria or endosymbionts of the algae in the culture medium (Supplemental Fig. S1 http://dx.doi.org/10.1017/S0954102014000017). The two genes are 31.73% identical at the amino acid level and are organized in tandem on the chromosome. Tandem organization of genes encoding AFPs has been reported in cold-adapted fishes (Scott et al. Reference Scott, Hayes, Fletcher and Davies1988, Wang et al. Reference Wang, DeVries and Cheng1995), but amongst bacteria it appears to be unique to this symbiont.
The relatively low sequence identity and the tandem organization suggest the occurrence of HGT between two bacterial species. The phenomenon of HGT between bacteria, and from bacteria to eukaryotes, is well documented; HGT frequently occurs in cases of selective pressure from harsh environmental conditions and is favoured by endosymbiosis (Sorhannus Reference Sorhannus2011). Sorhannus (Reference Sorhannus2011) reported four examples of HGT of AFP genes: one from Polaribacter irgensii to a group of four proteobacterium species, one from the diatom Chaetocerus neogracile to the copepod Stephos Longipes, one from a basidiomycete to the diatoms Fragilariopsis curta and Fragilariopsis cylindrus, and one from an ascomycete lineage to the Antarctic bacterium S. aurantiaca. The latter constitutes a rare example of HGT from eukaryotes to prokaryotes (Keeling & Palmer Reference Keeling and Palmer2008); however, this gene transfer event is disputed because neither of the ascomycete species is known to occur in polar regions (Sorhannus Reference Sorhannus2011). In our phylogenetic tree, the S. aurantiaca AFP appears to branch from the Antarctic basydiomycete G. antarctica, suggesting that S. aurantiaca acquired the AFP gene from basydiomycetes. Furthermore, our phylogenetic analysis showed that both EFsymbAFP and EFsymbIBP cluster principally with homologues from other Antarctic bacteria. The high bootstrap values strongly suggest that the EFsymbAFP and EFsymbIBP genes came from S. aurantiaca and F. bacterium lineages. However, it is possible that these sequences derived from close relatives of S. aurantiaca and F. bacterium that were not included in the current sample. Thus, the presence of the two genes in the E. focardii genome can be explained by two separate symbiotic events, after which the two genes were retained by E. focardii due to their high adaptive significance in cold environments.
From the modelling analysis, EFsymbAFP and EFsymbIBP appear similar to the AFPs from the fungus T. ishikariensis and from the Arctic yeast Leucosporidium sp. AY30. Both N- and C-terminal domains of EFsymbIBP are similar to the structure of the AFP from the fungi (Fig. 4); this supports the hypothesis that EFsymbIBP resulted from a gene duplication event. This gene organization is also present in F. bacterium (Raymond et al. Reference Raymond, Christner and Schuster2008), therefore the gene duplication most probably happened prior to the symbiotic relationship between the bacterium and E. focardii.
The EFsymbAFP and EFsymbIBP IBSs appear to maintain a relatively high content of hydrophobic and hydrogen-bond forming residues; these features are shared with other AFPs and help bind ice crystals through the ‘anchored clathrate’ mode of action, i.e. through the contributions of both hydrophobic effects and hydrogen bonds (Garnham et al. Reference Garnham, Campbell and Davies2011).
Genes coding for proteins with antifreeze properties probably evolved independently in a number of lineages, while other organisms acquired the genes through HGT (Kelley et al. Reference Kelley, Aagaard, MacCoss and Swanson2010). The evolution of the prokaryotic genome is thought to have been profoundly influenced by HGT (Keeling & Palmer Reference Keeling and Palmer2008). Most of the AFPs are derived from gene duplication events and from diversification of pre-existing genes that perform other functions. For example, the type II AFPs come from the duplication and diversification of the C-type lectin genes, and the Antarctic Notothenoid AFP is derived from a trypsinogen-like serine protease. This raises an interesting question: where did the bacterial AFP come from? Most of these bacterial sequences share a conserved domain (DUF3494), the function of which is unknown, but that may be considered as a fingerprint for IBPs. However, EFsymbAFP, EFsymbIBP and their closest homologues share a sequence similarity that ranges from 42% to 52% with the alpha 5 type IV collagen isoform 3 from Rhodococcus sp. JVH1. It has been reported that collagen peptides derived from the alcalase hydrolysis of bovine gelatine are able to inhibit the re-crystallization of ice in ice cream (Wang & Damodaran Reference Wang and Damodaran2009), suggesting that a bacterial collagen-like protein may represent the ancestor of the bacterial AFPs.
To conclude, our results confirm that IBPs and AFPs have complex evolutionary histories, which includes gene/domain duplication and HGT events, most probably due to the demands of the environment and the need for rapid adaptation.
Acknowledgements
This research was supported by grants from the Italian Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) (PRIN 2008) and the Italian Programma Nazionale di Ricerche in Antartide (PNRA). We acknowledge support from the Italian Ministry of Education and Research through the Flagship (PB05) InterOmics, HIRMA (RBAP11YS7K) and the European MIMOMICS projects. We are grateful to Vadim Gladishev and all the members of his laboratory for their collaboration in the sequencing the E. focardii genome. Special thanks to Alexei Lobanov for the helpful suggestions during the editing of the manuscript. We acknowledge the COST (action BM1102) for supporting part of this work. We also thank the reviewers for their comments.










